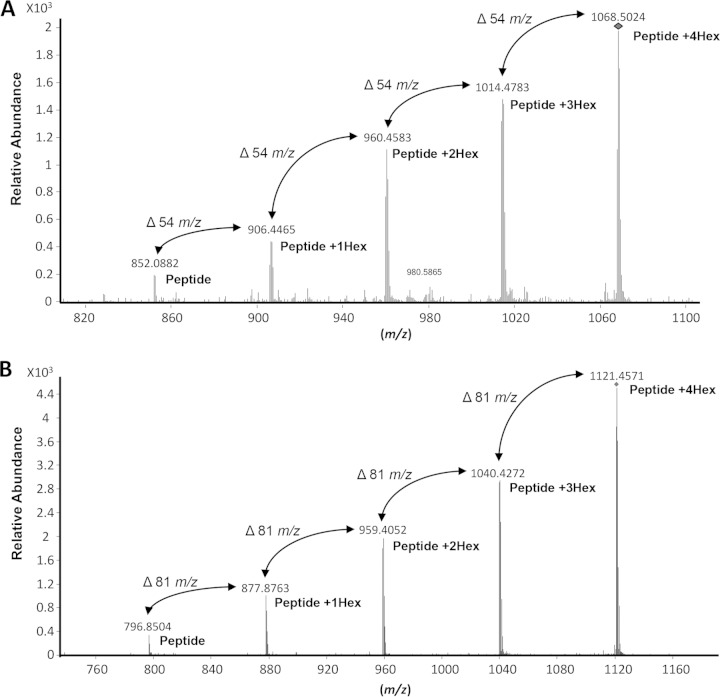
FIG 5.
Analysis of peptides derived from rSodC demonstrates protein glycosylation. (A) MS/MS analysis of glycopeptide (51TGSPAPSGMLGAEAESMGPPNIITRL76) using low collision energy. MS/MS data were collected using ramped collision energies, with the slope of the collision energy set to 3.0 V and an offset of 2.5 V. All MS/MS data were analyzed using Mascot with the following parameters: a parent ion tolerance of 2.5 Da, a fragment mass tolerance of 1.0 Da, up to two missed cleavages allowed, and variable Met oxidation. The MS/MS was analyzed for the neutral loss of m/z of 162.0528 using Mass Hunter Qualitative Analysis software, version B.04.00 (Agilent Technologies). MS/MS fragmentation identified the five most intense [M + 3H]3+ ion peaks. These molecular ions (m/z 852.0882, 906.4465, 960.4583, 1,014.4783, and 1,068.5024) differed by 54 m/z, corresponding to hexose residues of a triply charged mass of the glycopeptide. (B) MS/MS analysis of glycopeptide (51TGSPAPSGMLGAEAESM67) using low collision energy. MS/MS fragmentation identified the five most intense [M + 2H]2+ ion peaks. These molecular ions (m/z 796.8504, 877.8763, 959.4052, 1,040.4272, and 1,121.4571) differed by 81 m/z corresponding hexose residues of a doubly charged mass of the glycopeptide. The 1,068.5024 and 1,121.4571 m/z ions of the respective glycopeptides were subjected to de novo sequence analysis to confirm identity of the glycopeptide using high-collision-energy fragmentation (Fig. 6).