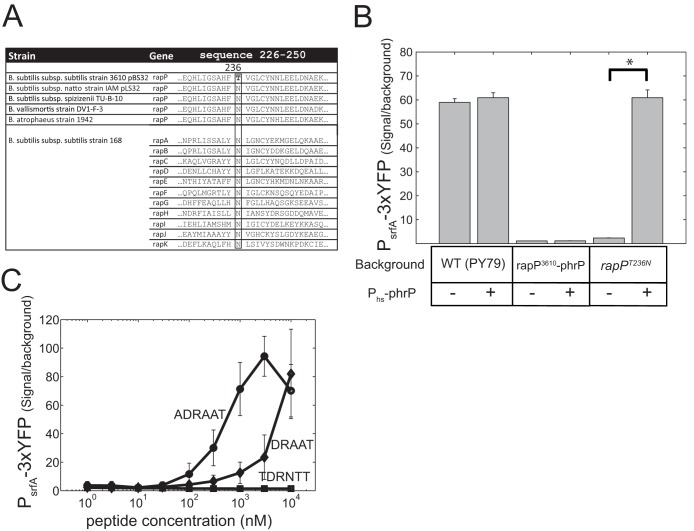
FIG 5.
rapP3610 codes for an N236T substitution mutation compared to the consensus, which renders it insensitive to the PhrP peptide. (A) Sequence alignment of amino acids 226 to 250 from RapP3610 with the sequences of other RapP homologues and with other Rap proteins found in strain 3610. Asparagine residue 236 is conserved in all Rap proteins except for the product of rapP3610, where it is replaced with threonine. Residue 236 of RapP is shown in boldface, and the aligned column is boxed for emphasis. (B) The addition of an IPTG-inducible phrP gene suppresses the effect of rapPT236N but not of rapP3610 on PsrfA-3×YFP expression. Shown are YFP expression levels of a PsrfA-3×YFP reporter in strain PY79 with different backgrounds, where each genotype was measured with (+) or without (−) a Phs-phrP construct. Genotypes are as follows: wild-type PY79 (AES1334), Phs-phrP (AES1477), PrapP-rapP3610 (AES1380), PrapP-rapP3610 Phs-phrP (AES1478), PrapP-rapPT236N (AES1678), and PrapP-rapPT236N Phs-phrP (AES1709). Expression was measured at an OD600 of 1.5, and IPTG was added at 10 μM when needed. (C) PsrfA-3×YFP expression was monitored in a PrapP-rapPT236N (AES1678) background as a function of the concentration of the externally added peptides. Three peptides were tested for their ability to repress rapPT236N activity: the putative signaling peptides of PhrP (DRAAT or ADRAAT) and the hexapeptide signal of PhrH (TDRNTT).