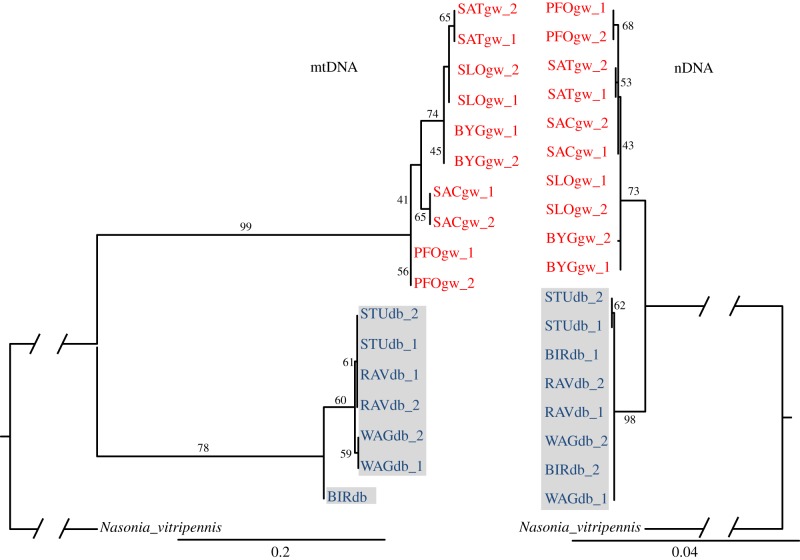
Figure 4.
Maximum-likelihood phylogenetic relationships of strains of Lariophagus distinguendus inferred from analyses of the partial mitochondrial gene COI (mtDNA) and combined sequences from the nuclear markers CAD, LOC100123206, LOC100123909, LOC100117339 and ITS2 (nDNA). COI was partitioned in first/second and third codon positions (electronic supplementary material, table S5). The nDNA dataset was partitioned into first/second codon positions (p1) and third codon positions (p2) when referring to exonic nucleotides, the intronic nucleotides of CAD and the LOC markers (p3), and ITS2 (p4; electronic supplementary material, table S5). Branch support values are given in percentages for maximum-likelihood bootstrapping (1000 bootstrap replicates). Light grey (red online): strains collected from drugstore beetles in private homes. Dark grey (blue online) with grey shading: strains collected from granary weevils in grain stores. (Online version in colour.)