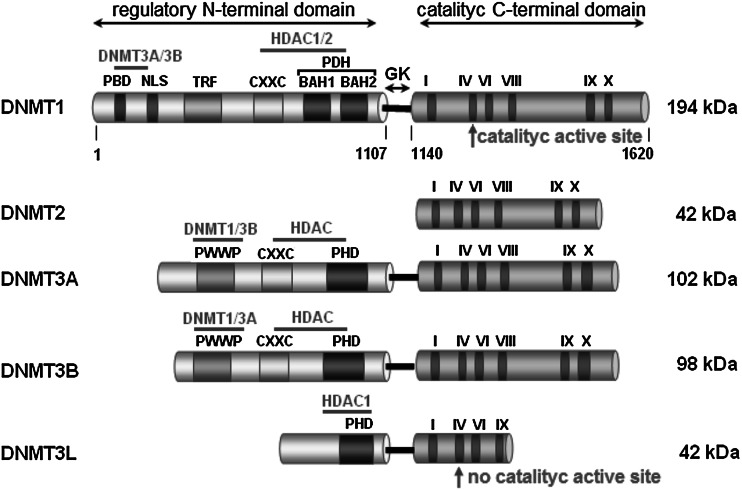
Fig. 1.
Schematic structure of mammalian DNMT family members. DNMT1, the first described methyltransferase, preferentially methylates hemimethylated DNA (Robertson 2001). DNMT2 lacks the N-terminal domain, while C-terminal domain contains the full set of sequence motifs but has not been shown to have transmethylase activity (Bestor 2000). DNMT3A and DNMT3B have similar domain arrangements and an equal preference for hemimethylated and unmethylated DNA (Robertson 2001). DNMT3L, being closely related to the catalytic domain of DNMT3A/3B, lacks canonical DNA cytosine−methyltransferase motifs (Bestor 2000). Its N-terminal regulatory domains exhibit little similarity but the catalytic domains of DNMTs are conserved. The N-terminal domain possesses: PBD—proliferating cell nuclear antigen (PCNA) binding domain, NLS—nuclear localization signal, TRF—targeting replication foci, CXXC—cysteine rich, zinc finger DNA-binding motif, PBH—polybromo homology domain, PWWP—tetrapeptide domain containing proline−tryptophan−tryptophan−proline motif. The N-terminal and C-terminal domains are linked by dinucleotide repeats: GK—glycine−lysine repeat. The C-terminal domain consists of six most conserved amino acid motifs (motif I and X form SAM binding site, motif IV binds cytosine at the active site). Mapped interaction sites of DNMTs and HDACs (histone deacetylases) are indicated in the above diagrams. The borders of the DNMT1 domains are marked according Song et al. (2011)