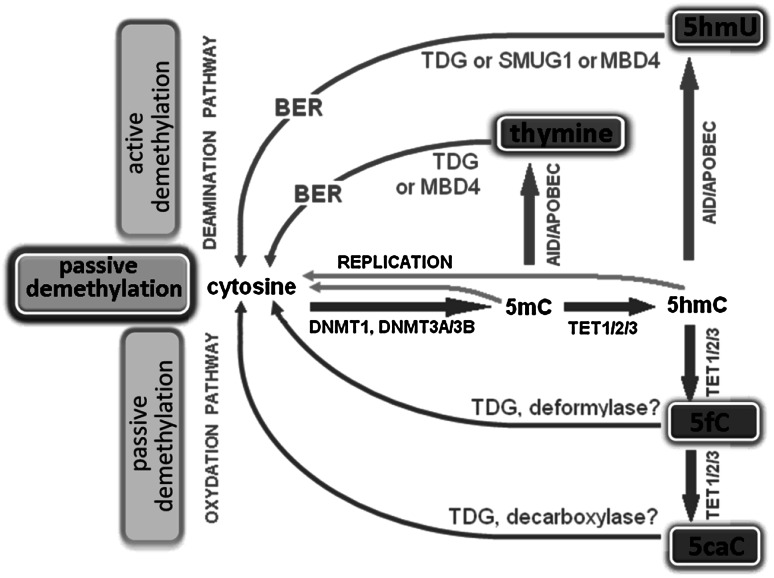
Fig. 2.
Passive and active DNA demethylation pathways. Passive DNA demethylation is caused by a reduction in activity or absence of DNMTs (yellow arrows). Active demethylation via oxidation pathway (green arrows): TET enzymes can hydroxylate methylcytosine (5mC) to form 5-hydroxymethylcytosine (5hmC); further oxidation produces 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). 5fC and 5caC can be actively removed by the DNA glycosylases. In addition, a putative deformylase may convert 5fC to C and decarboxylase convert 5caC to C. Active demethylation via deamination pathway (red arrows): AID/APOBEC family members can deaminate 5mC or 5hmC to form thymidine or 5-hydroxymethyluracil (5hmU). These intermediates are replaced by cytosine during base excision repair (BER) mediated by the uracil-DNA glycosylase (UDG) family, like TDG or SMUG1 as well as MBD4 (specifically recognize thymine and 5hmU). AID activation-induced deaminase, APOBEC apolipoprotein B mRNA-editing enzyme complex, BER—base excision repair, DNMT1/3A/3B—DNA methyltransferase, MBD4—methyl-binding domain protein 4, SMUG1—single-strand specific monofunctional uracil-DNA glycosylase, TET1/2/3—ten-eleven methylcytosine dioxygenase family, TDG—thymine-DNA glycosylase (Color figure online)