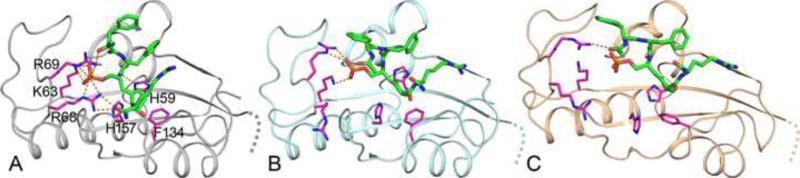
Figure 6. Docked poses for the trans ligand at the catalytic site, generated using RosettaLigand on conformations from our simulations.
(A) The optimal pose of the ligand, refined from the simulation of Pin1 with this ligand bound at the catalytic site (as well as the WW site). The ligand Pro residue is properly positioned relative to Pin1 His59, Phe134, and His157; the ligand pSer sidechain forms salt bridges with Lys63, Arg68, and Arg69 of the catalytic loop.
(B) The ligand docked to the empty catalytic site of a conformation from the simulation of Pin1 with FFpSPR bound at the WW site. The ligand Pro residue is also properly positioned, and the pSer sidechain still forms some of the salt bridges in (A).
(C) The ligand docked to the catalytic site of a conformation from the simulation of apo Pin1. The ligand Pro residue is shifted to the right, and the pSer sidechain loses all the salt bridges.
See also Figure S10.