Fig. 4.
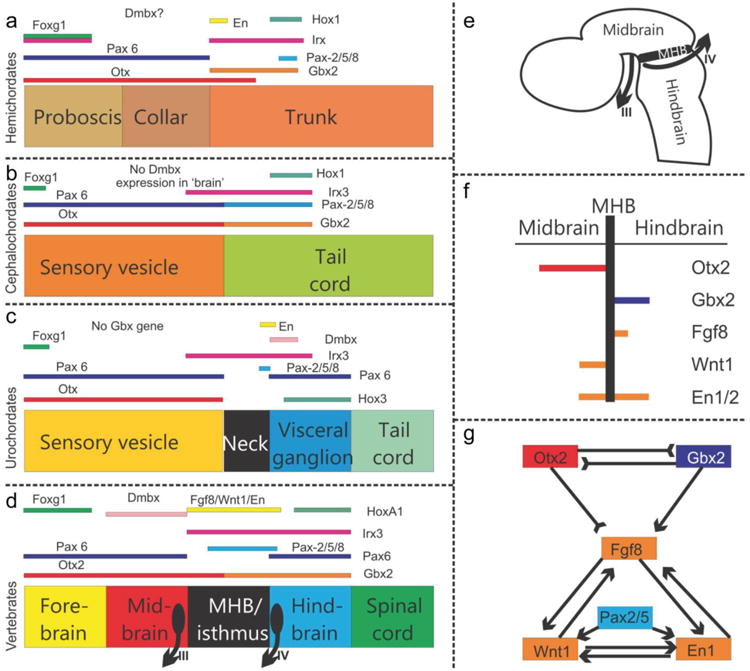
The evolution of gene expression at the midbrain–hindbrain boundary (MHB) is shown for deuterostomes. The MHB of vertebrates shows abutting Otx2 and Gbx2 expression (D-G). This stabilizes the expression of Fgf8 (G), which in turn stabilizes the expression of Wnt1 and engrailed (En1). Mutation of Otx2, Gbx2, Fgf8, or Wnt1 eliminates the MHB. Pax2/5/8 are also expressed at the MHB, whereas the expression of Dmbx occurs immediately rostral to the MHB in the midbrain to later expand into the hindbrain and spinal cord (D). Note the partial overlap of Pax2/5/8 with the caudal expression of Otx2 and the rostral expression of Gbx2 (D). Hemichordates (A) have overlapping expression of Gbx, Otx, Irx and En in the rostral trunk. Pax6 abuts Gbx2 whereas Pax2/5/8 overlaps with the caudal expression of Gbx2. Outgroup data suggest that coelenterates have a Dmbx ortholog, thus raising the possibility that hemichordates (A) also have a Dmbx gene. Cephalochordates (B) have no Dmbx expression in the ‘brain’. Otx abuts with Gbx, like in vertebrates. However, Gbx overlaps with Pax2/5/8 and most of Irx3. Urochordates (C) have no Gbx gene but have a Pax2/5/8 and Pax6 configuration comparable to vertebrates. Dmbx overlaps with the caudal end of the Irx3 expression whereas Dmbx expression is rostral to Irx3 in vertebrates. Together these data show that certain gene expression domains are topographically conserved (Foxg1, Hox, Otx), whereas others show varying degrees of overlap. It is conceivable that the evolution of nested expression domains of transcription factors is causally related to the evolution of specific neuronal features such as the evolution of oculomotor and trochlear motoneurons (D,E) around the MHB. Experimental work has demonstrated that the development of these motor centers depends on the formation of the MHB. Adopted from (Beccari, et al., 2013, Fritzsch and Glover, 2006, Pani, et al., 2012)
