Abstract
Understanding the genes and mechanisms involved in acute alcohol responses has the potential to allow us to predict an individual’s predisposition to developing an alcohol use disorder. To better understand the molecular pathways involved in the activating effects of alcohol and the acute functional tolerance that can develop to such effects, we characterized a novel ethanol-induced hypercontraction response displayed by Caenorhabditis elegans. We compared body size of animals prior to and during ethanol treatment and showed that acute exposure to ethanol produced a concentration-dependent decrease in size followed by recovery to their untreated size by 40 min despite continuous treatment. An increase in cholinergic signaling, leading to muscle hypercontraction, is implicated in this effect because pretreatment with mecamylamine, a nicotinic acetylcholine receptor (nAChR) antagonist, blocked ethanol-induced hypercontraction, as did mutations causing defects in cholinergic signaling (cha-1 and unc-17). Analysis of mutations affecting specific subunits of nAChRs excluded a role for the ACR-2R, the ACR-16R, and the levamisole-sensitive AChR and indicated that this excitation effect is dependent on an uncharacterized nAChR that contains the UNC-63 α-subunit. We performed a forward genetic screen and identified eg200, a mutation that affects a conserved glycine in EAT-6, the α-subunit of the Na+/K+ ATPase. The eat-6(eg200) mutant fails to develop tolerance to ethanol-induced hypercontraction and remains contracted for at least 3 hr of continuous ethanol exposure. These data suggest that cholinergic signaling through a specific α-subunit-containing nAChR is involved in ethanol-induced excitation and that tolerance to this ethanol effect is modulated by Na+/K+ ATPase function.
Keywords: ethanol, Caenorhabditis elegans, nicotinic acetylcholine receptors, excitation, tolerance
THE abuse of alcohol is a cause of significant societal and health-related problems. Despite the common usage of this drug, the molecular underpinnings of alcohol’s acute actions on the brain are poorly understood. Alcohol (ethanol) has biphasic behavioral effects in humans and other animals, acting as a stimulant at lower concentrations and as a depressant at higher concentrations. Understanding the molecular nature of these biphasic effects is made difficult by the fact that ethanol interacts with, and alters the function of, many proteins, including neurotransmitter receptors and ion channels (Harris et al. 2008; Spanagel 2009). A commonly suggested class of ethanol targets is ligand-gated ion channels (LGICs), which include NMDA glutamate receptors, and members of a subclass of LGICs, the Cys-loop superfamily, including GABAA, glycine, and nicotinic acetylcholine receptors (nAChRs) (Olsen et al. 2014). Effects on the GABAA and glutamate receptors are hypothesized to play a significant role in the depressant effects of ethanol (Harris et al. 2008). Locomotor activation by low to moderate concentrations of ethanol in rodents is thought to model the euphoric effects of ethanol in humans (Phillips and Shen 1996). Locomotor activation by ethanol has been linked to increased dopamine release in neural reward pathways, possibly via mechanisms that involve inhibition of the inhibitory GABAB metabotropic receptors by ethanol (Holstein et al. 2009; Kruse et al. 2012) and/or by ethanol activation of neuronal nAChRs (Soderpalm et al. 2000; Larsson et al. 2002; Kamens and Phillips 2008; Kamens et al. 2009).
Individual variation that alters the mechanisms responsible for the acute behavioral effects of ethanol is likely to contribute to variation in predisposition to develop alcohol use disorders (AUDs). An individual’s level of response (LR) to acute ethanol intoxication is determined by two major opposing biological forces, the degree of sensitivity to intoxication and the rate at which tolerance develops to that acute intoxication during the intoxication session (called acute functional tolerance) (Newlin and Thomson 1990). LR has genetic determinants (Kalu et al. 2012) and LR in naive drinkers is a predictor of the predisposition to develop AUDs later in life (Schuckit 1994; Volavka et al. 1996; Heath et al. 1999). Therefore, it is important that we better understand the pathways responsible for the behavioral actions of ethanol and the mechanisms by which acute functional tolerance can develop to those behavioral effects. Studies in selectively bred mouse strains suggest that ethanol-induced locomotor activation and the rate of development of acute functional tolerance may share common mechanisms (Ponomarev and Crabbe 2002).
Caenorhabditis elegans represents a powerful model for identifying drug targets, using genetic approaches (Van Swinderen et al. 1999; Davies et al. 2003; Artal-Sanz et al. 2006; Kaletta and Hengartner 2006; Kwok et al. 2006). C. elegans are sensitive to acute ethanol exposure, demonstrating an increased speed of locomotion at low ethanol concentrations (Graham et al. 2009) and decreased amplitude of body bends and decreased rates of crawling, swimming, egg laying, and pharyngeal pumping at higher concentrations of ethanol (Davies et al. 2003; Mitchell et al. 2007; Speca et al. 2010). Ethanol also produces disinhibition of certain crawling behaviors that would normally be inhibited in a liquid environment (Topper et al. 2014). Acute functional tolerance, which is unrelated to metabolism of the drug, develops rapidly to the effects of ethanol on the speed of locomotion (Davies et al. 2004; Alaimo et al. 2012; Bettinger et al. 2012; Raabe et al. 2014). Longer ethanol exposures followed by removal from the drug can lead to withdrawal-related phenotypes (Davies et al. 2004; Mitchell et al. 2010). Therefore, these drug-induced behavioral effects are indicative of ethanol having activating and depressing actions in C. elegans as it does in mammals and demonstrate that tolerance can develop to acute and long-term ethanol exposures.
In this study, we describe a novel excitation effect of acute ethanol treatment of C. elegans, which we have termed ethanol-induced hypercontraction (EHC). EHC is time dependent; treated animals show a rapid onset of body hypercontraction and a relatively quick recovery while in the presence of the drug. This latter observation is indicative of the development of acute functional tolerance. We test the role of candidate neurotransmitter signaling systems in EHC and identify a specific nAChR subunit that mediates the excitation effect. We describe the identification of a mutant that displays a lack of development of tolerance to EHC as an allele of eat-6, the Na+/K+-ATPase α-subunit-encoding gene, suggesting the possibility that Na+/K+-ATPase function is required for the development of tolerance to EHC.
Materials and Methods
Nematode maintenance and strains
Nematodes were maintained at 20° on nematode growth medium (NGM) plates in the presence of Escherichia coli strain OP50. The strains used in this study were N2 (wild type), CB4856 (wild type), eri-1(mg366), cha-1(p1152), unc-17(e245), unc-17(e113), acr-2(ok1887), acr-16(ok789), unc-38(e264), unc-29(e193), lev-1(e211), unc-63(x13), unc-63(gk234), gbb-1(tm1406), gbb-2(tm1165), unc-25(e156), unc-25(sa94), unc-25(n2569), unc-49(e407), unc-30(e191), unc-30(e318), unc-47(gk192), unc-47(e307), slo-1(eg142), npr-1(ky13), eat-6(eg200), eat-6(ad601), eat-6(ad467), eat-6(ad997), egl-3(n150), unc-31(e928), nlp-12(ok335), trp-4(sy695), lon-3(e2175), unc-61(e228), unc-42(e270) yDf12 V/nT1[unc-?(n754) let-?](IV;V); dpy-6(e14)X, unc-42(e270) arDf1 V/nT1[unc-?(n754) let-?](IV;V), and nDf42 V/nT1[unc-?(n754) let-?](IV;V). Strains were provided by the Caenorhabditis Genetics Center (CGC) and the National Bioresource Project for the nematode (Japan).
Hypercontraction assay
Plates were prepared as previously described (Davies et al. 2003). Briefly, NGM plates (6 cm) were dried for 1.5 hr at 37° 18–20 hr before experiments were performed. Plates were weighed (to determine media volume) and four copper rings were melted into the surface of the agar of each plate to corral the worms. Plates were divided into the following groups: (1) acclimation, (2) 0 mM ethanol treatment, and (3) X mM ethanol treatment, where X represents the final exogenous concentration of ethanol (from 100 to 500 mM). Ethanol was added to the ethanol treatment plates (vol/vol) to the desired concentration and allowed to equilibrate for 2 hr before the experiment was performed. Ten age-matched first-day adults were assayed in each of the four rings. The acclimation plates were used to acclimate the worms to the absence of bacteria (food) for 30 min prior to the experiment being performed. The worms were then transferred to their respective treatment plates for the assay (0 mM or X mM ethanol). We have previously shown that a 400-mM exogenous dose generates a tissue concentration of ethanol in C. elegans of ∼44 mM (Alaimo et al. 2012).
To measure worm perimeter, images were captured with a Retiga 4000R (QImaging, BC, Canada) camera attached to an Olympus SZX7 microscope set to a magnification of 2.0× and fitted with a 0.5× objective. Images were taken using ImagePro Plus Version 6 (Media Cybernetic, Bethesda, MD) at the following time points: −1 min (on the acclimation plate, immediately prior to movement to the treatment plate) and 1, 5, 10, 20, 30, and 40 min. Worm size was determined using a macro written in the ImagePro Plus program (available on request). The macro converted the captured images to 8-bit gray scale and filtered the images to enhance the worm objects. First, the image was flattened (feature width 20) and then a large spectral filter was applied (Edge Minus, W10, H10, S1, P1). The resulting image was inverted and the count/size feature was applied to detect the worm objects based on a gray-scale threshold. Most nonworm objects were excluded based on a size filter, using parameters that include area, feret size, and perimeter. Finally, a manual visual check was used to ensure that only worms were included in the measurements and that no worms were included that had generated a ring shape, which can form during an omega bend movement. The average perimeter size for the group of 10 animals (n = 1) in each copper ring was determined at each time point. To control for differences in the baseline perimeter for strains of different genotypes (Supporting Information, Table S2), data are presented as perimeter (% of untreated) ± SEM and were calculated using the following equation:
With the exception of Figure 1B, the data for worms assayed in the absence of ethanol are not shown. In the absence of ethanol, we found no significant differences for perimeter size measured over time for any of the strains we tested.
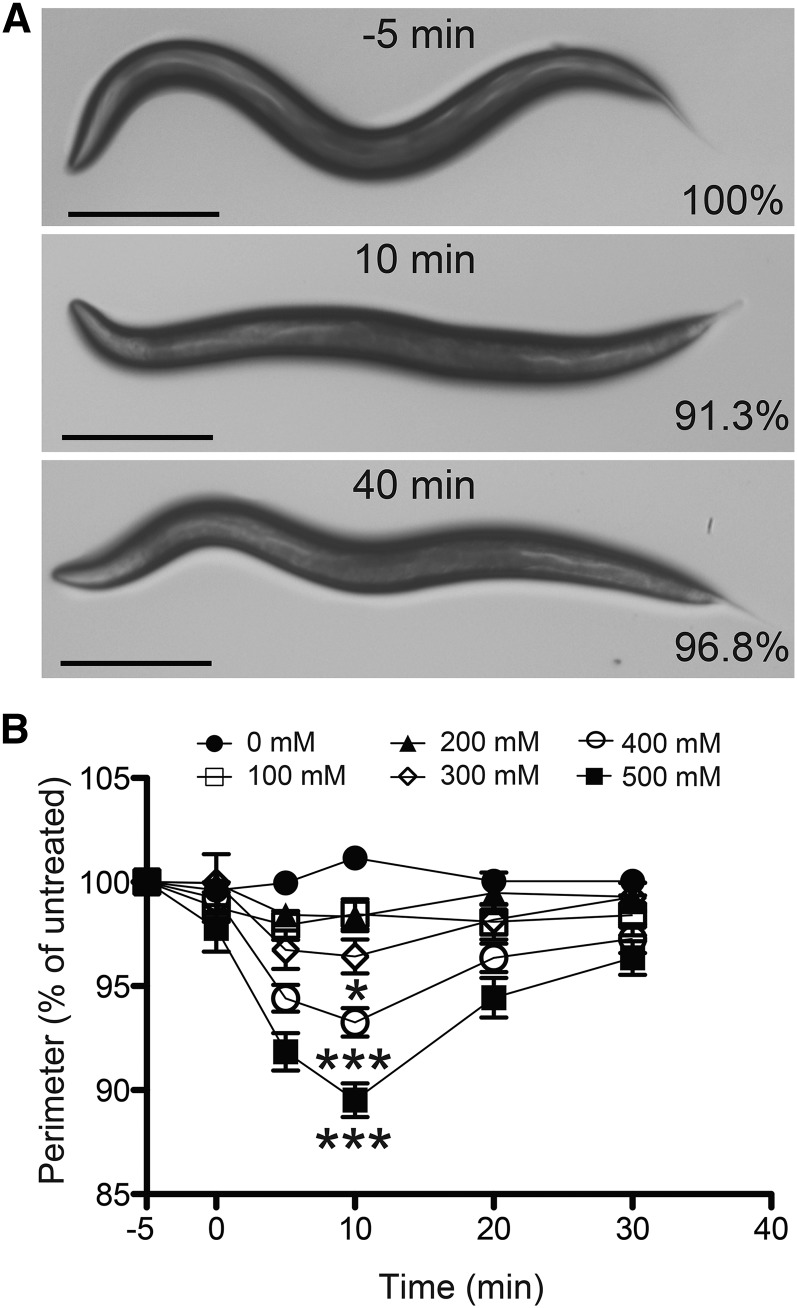
Figure 1.
Ethanol dose-dependently induces hypercontraction. (A) Images of the same N2 (wild-type) worm before treatment (−5 min) and during treatment with 400 mM ethanol (10 and 40 min of exposure). Percentage indicates percentage of untreated perimeter at −5 min. Bar, 250 μm. (B) N2 (wild-type) worms treated with 0, 100, 200, 300, 400, and 500 mM exogenous ethanol. A significant decrease in size, indicating contraction, relative to 0 mM was obtained at the 10-min time point at 300, 400, and 500 mM ethanol. Recovery from the contraction was significant at 30 min at concentrations of 400 and 500 mM ethanol (*P < 0.05; ***P < 0.001; n = 12).
Locomotion
Basal speeds were determined as previously described (Alaimo et al. 2012). The plates and first-day adult animals were prepared as described for the hypercontraction assay. A 2-min movie was recorded at 30 min and the average speed was measured using ImagePro Plus Version 6 (Media Cybernetics, Bethesda, MD). The average speed and SEM of six trials were plotted and analyzed using a one-way ANOVA with Prism v 5.0 (GraphPad Software).
Identification of the eg200 mutation
The eg200 mutant was isolated in a small forward genetic screen (1800 haploid genomes tested on 150–200 mM ethanol) on the basis of slower than wild-type speeds in the presence of ethanol. The prolonged ethanol-induced hypercontraction phenotype associated with eg200 was used thereafter in the mapping and identification of the causative mutation. Using SNP mapping methods (Wicks et al. 2001; Davis et al. 2005) with the Hawaiian wild-type strain CB4856, the eg200 mutation was initially mapped to the right arm of chromosome V between two SNPs ∼1.8 map units apart (pkP5068 and pkP5087). The SNPs assessed in fine-mapping eg200 on chromosome V were haw78359, pkP5067, pkP5068, pkP5069, pkP5125, pkP5087, pkP5086, pkP5084, and pkP5126 (http://wormbase.org). To facilitate the recovery of additional recombinants in the genetic interval containing eg200, a triple-mutant strain, lon-3(e2175) eg200unc-61(e228), was constructed using standard genetic crosses. The triple mutant was crossed to CB4856, F1 progeny were allowed to self-fertilize, and F2 progeny were isolated that were Lon non-Unc or Unc non-Lon. The recombinant chromosome was made homozygous by selecting self-progeny individuals that were Lon non-Unc or Unc non-Lon and did not have self-progeny that were Lon Unc. The presence or absence of the eg200 mutation in these recombinant strains was determined based on the EHC phenotype. The strains were then examined for N2/CB4856 SNPs in the interval. This mapping further narrowed the eg200-containing interval to between pkP5068 and snp_F57B1[3] (http://wormbase.org).
The eg200 mutation failed to complement the deficiencies yDf12 and nDf42, while complementing arDf1. To do this, eg200 homozygous males were crossed to the deficiency strains and non-Unc (eg200/Df) young adult progeny were tested for prolonged contraction in the presence of ethanol. Both yDf12 and arDf1 are known to delete the lin-25 gene (Hsu and Meyer 1994; Tuck and Greenwald 1995), while the max-1 gene is deleted by yDf12 but not by arDf1 (Huang et al. 2002), which suggested that eg200 was in the vicinity of max-1 to the right of the arDf1 right breakpoint. To determine the extent of the right breakpoints of arDf1 and yDf12, the deficiency-bearing strains were crossed to CB4856 males and DNA was prepared from non-Unc (+/Df) progeny. The primers used to amplify DNA fragments (i–viii, shown in Figure 6D) in the interval of interest were previously described (Swan et al. 2002) (File S1, Table M1). Each of these primer pairs flanks a known dimorphic SNP present in the N2 and CB4856 strains. Sequencing of the amplified fragment was carried out to determine the genotype (only the CB4856 allele would be detected if the region is deleted by the deficiency or both the N2 and CB4856 alleles would be detected if the interval is not deleted by the deficiency). These experiments placed the right breakpoint of arDf1 between lin-25 and the PCR fragment labeled i in Figure 6D, whereas the right breakpoint of yDf12 was between PCR fragments vii and viii.
Figure 6.
Mutations in the eat-6 gene alter tolerance to ethanol-induced hypercontraction. (A) The eg200 mutant animals have decreased movement compared with wild-type animals on ethanol. In these representative illustrations, images of wild-type and eg200 worms were captured every 10 sec over a 2-min period after 30 min of ethanol exposure (400 mM). The progression of time is indicated by darkening worm silhouettes. Bars, 2 mm. (B) Average speeds of wild-type and eg200 mutant animals in the absence and presence of ethanol (400 mM) after 30 min of exposure (***P < 0.001; n = 5). (C) The eat-6(eg200) mutation increases the sensitivity to paralysis caused by prolonged exposure to the AChR agonist, levamisole (100 μM) (**P < 0.01; ***P < 0.001; n = 3). (D) The eg200 mutation was mapped between SNPs on chromosome V and between endpoints of the deletions arDf1 and yDf12. The approximate endpoints of the deletions were determined by placing CB4856-specific SNPs (i–viii) in trans to the deletion-bearing chromosomes and sequencing to determine genotype at each SNP (hemizygous CB4856 or heterozygous N2/CB4856). (E) The eg200 mutation is predicted to cause a substitution of a glutamic acid for a conserved glycine (Gly77) in the EAT-6 protein. The mutation is a G to A substitution in exon 2 of eat-6 (nucleotide 230 of the mRNA sequence; position 13,129,568 of the chromosome V nucleotide sequence).
We chose eat-6 as a candidate gene within the eg200-containing genetic interval to test. To confirm that eat-6 was affected by the eg200 mutation, the genomic region spanning eat-6 coding sequences was amplified by PCR, using genomic DNA purified from the eg200 mutant as the template for the reaction (File S1, Table M2).
Aldicarb and levamisole treatment
The aldicarb assay was adapted from previously described methods (Mahoney et al. 2006). Aldicarb (Sigma-Aldrich, St. Louis) plates were prepared at a concentration of 0.5 or 1 mM 2–3 days before the assay. For assays determining the sensitivity to aldicarb-induced paralysis, 10 first-day adults were moved to the aldicarb treatment plates (1.0 mM) and animals were scored for paralysis every 30 min for 3 hr. Worms were counted as paralyzed if they did not move when prodded with a platinum pick on the head three times. For assays determining the degree of hypercontraction induced by aldicarb treatment, first-day adults were moved to the aldicarb plates (0.5 or 1.0 mM) after their body size was determined on an aldicarb-free plate, and then perimeters were measured as described above for hypercontraction assays. Levamisole paralysis assays were carried out as previously described (Nurrish et al. 1999). Age-matched young adult animals were transferred to plates with agar containing 100 µM levamisole (Sigma-Aldrich) and paralysis induced by the drug was determined (as above) at 0, 10, 30, 60, 90, and 120 min of exposure.
Mecamylamine treatment
First-day adult worms were pretreated with mecamylamine (Sigma-Aldrich) by constructing small-volume NGM pads containing 5 mM of the compound. Small-volume pads were used to reduce the quantity of drug required for treatment. Copper rings were set in the lid of 24-well plates. Agar from NGM plates dried as explained in the hypercontraction assay section was melted in a water bath at 85°. Mecamylamine was added to the melted NGM to 5 mM and 200 µl of this solution was pipetted into each copper ring and allowed to harden at room temperature. Equivalent 200-µl pads made without mecamylamine were used for acclimation, preexposure measurements, untreated controls, and ethanol treatment. Ethanol added to the ethanol-treatment pads was allowed to equilibrate for 2 hr prior to their use.
Statistical analysis
Treated mutant strains were compared to treated wild-type controls to determine whether the mutation caused a significant effect on the EHC phenotype. Respective time points were compared against one another, using a repeated-measures two-way ANOVA with Bonferroni post hoc tests unless otherwise indicated (GraphPad Prism).
Results
The ethanol-induced hypercontraction phenotype is dose dependent
In performing assays of the behavioral effects of ethanol on C. elegans locomotion, we observed that acutely exposed worms demonstrate a transient decrease in overall body length, which we have termed EHC (Figure 1A). We performed a dose-response analysis to quantify the degree of hypercontraction and to determine the duration of this effect of ethanol on wild-type (WT) N2 young adult animals. Worms were exposed to 0-, 100-, 200-, 300-, 400-, and 500-mM exogenous concentrations of ethanol over a 30-min period and their lengths were measured and compared with their preexposure untreated lengths (Figure 1B). Higher concentrations (600 and 700 mM ethanol) were also tested but ethanol-induced coiling of the treated worms limited collection of the length measurements (Table S1), which may bias the data to overrepresent less-affected animals. Over the time course of exposure, there were two apparent phases in the EHC phenotype: (1) an initial contraction phase (0–10 min) and (2) a recovery phase (10–30 min). Both phases of the EHC phenotype are shown in Figure 1A, which contains images of the same N2 worm untreated (−5 min) and treated with 400 mM ethanol at 10 min and 40 min. Also apparent in Figure 1A, in the second image, is the decrease in body bend amplitude induced by ethanol (Davies et al. 2003). This flattening of the worm obscures the length decrease visually, but computer-assisted length measurements of midline or perimeter reveal the effect of ethanol on overall body length. The worms demonstrated a maximum contraction at 10 min with significant differences from the 0-mM untreated controls at 300, 400, and 500 mM ethanol (Figure 1B; Table S1). Significant recovery from EHC was achieved within 20 min following the point of maximum contraction (10 min) at both 400 and 500 mM ethanol. We chose to investigate EHC further using 400 mM ethanol, which was the lowest exogenous concentration at which we saw both statistically significant hypercontraction and a significant recovery from that hypercontraction.
Hypertonic-sensitive worms are not hypersensitive to ethanol
Hypertonic environments have previously been shown to produce a transient shrinkage in C. elegans (Choe and Strange 2007). We observed that hypertonic-induced hypercontraction differs substantially from EHC; hypertonic conditions (e.g., 349 mM NaCl) produce a greater decrease in body size and require a more prolonged recovery (4–5 hr) than hypercontraction of wild-type animals treated with 400 mM ethanol. We tested whether the mechanisms of EHC overlap with those responsible for the hypertonic hypercontraction response. Recovery from hypertonic shrinkage involves the activation of the with-no-lysine (WNK) WNK-1 serine–threonine kinase (Choe and Strange 2007). In agreement with this report, we found that RNA interference (RNAi) knockdown of WNK-1 inhibited recovery from hypertonic shock in eri-1(lf) worms (Figure S1A) [eri-1(mg363) animals are hypersensitive to RNAi]. If EHC occurred as a result of hypertonic stress, we hypothesized that knockdown of the wnk-1 gene should increase sensitivity to ethanol or prolong the ethanol effect. When we examined the sensitivity to EHC of wnk-1 RNAi-treated animals, we found that there was no difference in the degree of EHC compared to that in RNAi control-treated animals (Figure S1B), suggesting that the mechanisms of EHC do not overlap with the mediators of the response to hypertonic stress.
Other physical environmental properties of ethanol do not produce hypercontraction
We considered the possibility that other changes in the physical environment produced by ethanol might be the cause of EHC. At the concentration of ethanol we used, neither acidity nor viscosity are expected to be different from that in NGM media alone (Bates et al. 1963; Khattab et al. 2012). In the sealed petri plate, we might expect the presence of ethanol to alter vapor pressure. We tested vapor pressure, using a closed chamber system that we have used previously in the analysis of volatile inhalant drugs (Davies et al. 2012). Vapor pressure produced by 400 mM ethanol did not change the contraction state of wild-type animals (Figure S1C).
Hypercontraction to ethanol requires cholinergic signaling
Cholinergic signaling promotes contractile responses while GABAergic signaling induces relaxation responses in the longitudinal body wall muscle of the animal (Rand 2007). Pharmacological treatment of wild-type worms that leads to prolonged activation of cholinergic signaling has been shown to cause hypercontraction and paralyzed phenotypes (Lewis et al. 1980). While the worms displaying EHC are not paralyzed, we suspected that increased cholinergic signaling might be the mechanism responsible for EHC.
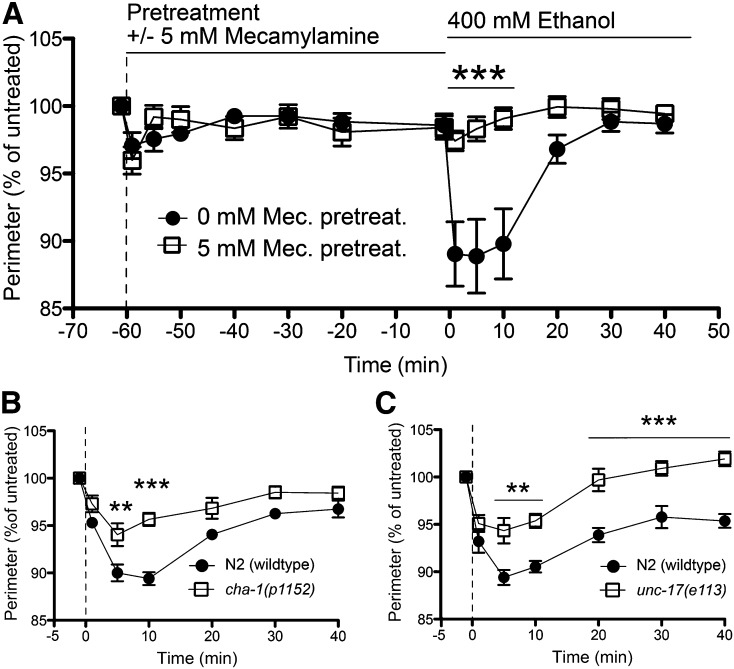
Mecamylamine is a nonselective noncompetitive antagonist of nAChRs. Preincubation of N2 worms for 1 hr with 5 mM mecamylamine did not significantly affect the body size of the worms during the pretreatment period (Figure 2A). When we pretreated animals with 5 mM mecamylamine and then exposed those worms to ethanol, we found that mecamylamine treatment significantly blocked EHC (Figure 2A). This result suggests that nAChR activity is required for ethanol to induce hypercontraction. We tested this hypothesis further, using mutants in genes that are involved in cholinergic signaling.
Figure 2.
Hypercontraction requires cholinergic signaling. (A) Preincubation (−60 to 0 min) with mecamylamine (Mec.) significantly blocked the EHC phenotype induced by 400 mM ethanol on N2 (wild-type) worms at 10 min (***P < 0.001; n = 11). (B and C) Mutant worms deficient in presynaptic acetylcholine release, cha-1 and unc-17, were significantly resistant to the hypercontraction induced by ethanol compared with N2 worms (**P < 0.01; ***P < 0.001; n = 6).
The synthesis of acetylcholine (ACh) requires the choline acetyltransferase (ChAT) enzyme that is encoded by the cha-1 gene (Rand and Russell 1984). Packaging into vesicles in presynaptic neurons requires the vesicular acetylcholine transporter (VAChT) that is encoded by the unc-17 gene (Alfonso et al. 1993). Strains carrying partial loss-of-function mutations in cha-1 or unc-17 have decreased levels of ACh released into the synaptic cleft (Rand 2007). When cha-1 or unc-17 mutant worms were tested on ethanol, we observed significant resistance to EHC compared to that in wild-type worms (Figure 2, B and C). The unc-17(e113) mutant showed an unusual tolerance response; by 40 min of exposure, the mutant worms recovered to a size greater than their preethanol treatment size (Figure 2C). A second allele, unc-17(e245), displayed the same resistance and tolerance phenotype (Table S2), confirming that this observation was not allele specific or a product of genetic background. The unc-17 and cha-1 data are in agreement with the mecamylamine data; together these results suggest a cholinergic mechanism of action for EHC.
Specific nAChRs play a role in EHC
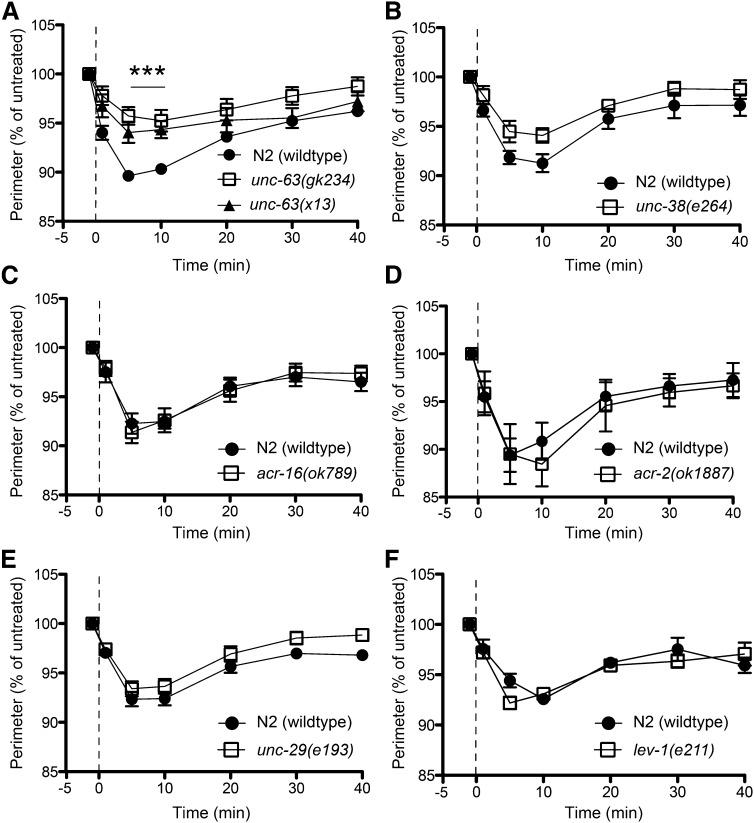
The C. elegans genome contains a large family of nAChRs with 29 identified subunits compared to the human genome, which contains 17 (Brown et al. 2006; Jones et al. 2007; Rand 2007). The most well-characterized subunits in worms are those expressed in motor neurons and at the neuromuscular junction (Richmond and Jorgensen 1999; Touroutine et al. 2005) and we tested many of these for a role in EHC. We focused first on the neuromuscular junction (NMJ) as EHC appears to involve muscle hyperactivation. Two receptor classes have been identified at the NMJ, those that are sensitive to the AChR agonist levamisole and those that are insensitive to levamisole. The levamisole-sensitive receptor requires the UNC-29, UNC-38, UNC-63, and LEV-1 subunits (Fleming et al. 1997; Culetto et al. 2004; Towers et al. 2005). The homomeric ACR-16R nAChR is the only defined levamisole-insensitive receptor at the NMJ (Touroutine et al. 2005). We tested members of each receptor class (levamisole sensitive, unc-29, unc-38, unc-63, and lev-1; levamisole insensitive, acr-16) for the effect of loss of function on EHC responses (Figure 3). We found that, of the genes tested, only mutations causing a loss of function in the unc-63 gene (x13 and gk234) significantly reduced the EHC response compared with that in wild-type animals (Figure 3A). The unc-38(e264) mutation produced a nonsignificant trend toward resistance to EHC (Figure 3B). That the unc-29 and lev-1 mutants displayed wild-type sensitivity to EHC (Figure 3, E and F) suggests that it is not the levamisole-sensitive receptor mediating the EHC effect because UNC-29 and LEV-1 are required subunits of that receptor. Instead, these data suggest the involvement of the UNC-63 subunit in a different receptor(s). One likely candidate was the neuronal ACR-2 receptor (ACR-2R), which has been shown to require UNC-63 and UNC-38 subunits (Jospin et al. 2009) and is expressed in cholinergic motorneurons (Petrash et al. 2013). Loss of function of the acr-2 gene eliminates the ACR-2R, so we tested acr-2(ok1887) for EHC, with the idea that if UNC-63 were acting in the ACR-2R to mediate EHC, then loss of ACR-2R should mimic the unc-63(lf) for this phenotype. acr-2(ok1887) mutants showed wild-type sensitivity to EHC (Figure 3D), which suggests that UNC-63 may belong to yet another nACh receptor class that is independent of the levamisole-sensitive receptor and the ACR-2R. These data indicate that the UNC-63 α-nAChR subunit plays a significant role in mediating EHC, and we have eliminated the levamisole-sensitive receptor and the ACR-2R as being required for the EHC effect.
Figure 3.
Ethanol-induced hypercontraction requires UNC-63. (A) Two different alleles of unc-63(gk234) and unc-63(x13) significantly reduced the hypercontraction response to 400 mM ethanol (***P < 0.001; n = 7). (B–F) unc-38 (n = 6), acr-16 (n = 6), acr-2 (n = 5), unc-29 (n = 10), and lev-1 (n = 5) mutants did not demonstrate a significant difference from N2 (wild type) (P > 0.05). Given the conserved homology between unc-63 and unc-38, the trend toward resistance for unc-38 may be relevant.
GABA signaling in EHC
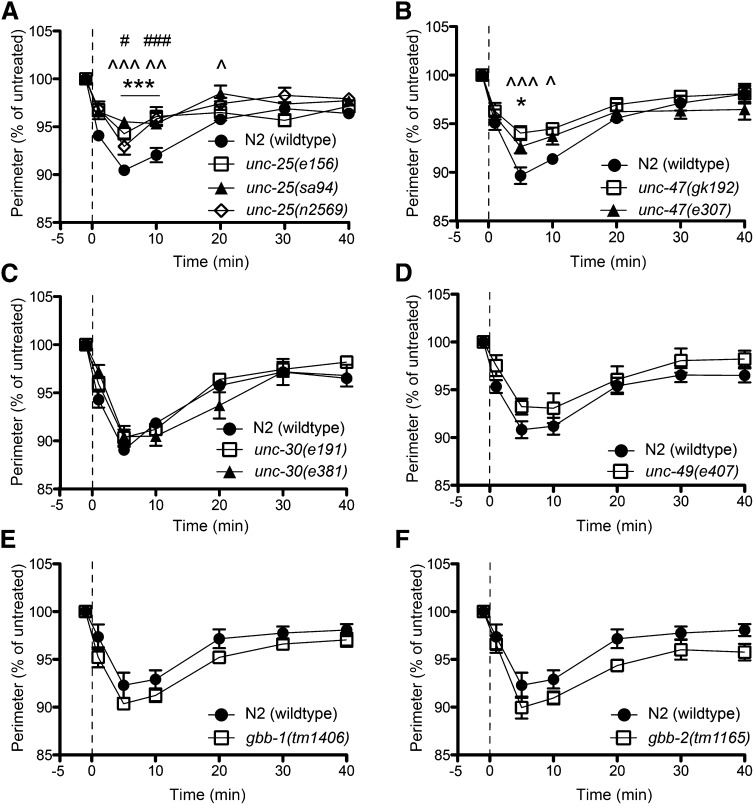
While ACh induces contraction of body wall muscle, GABA causes relaxation of the contralateral body wall muscle. We predicted that loss of GABA signaling might increase the degree of EHC due to a loss of inhibition at the NMJ. In contrast, we found that loss-of-function mutations in the unc-25 gene, which encodes the GABA synthetic enzyme, glutamic acid decarboxylase (GAD), decrease the degree of EHC (Figure 4A). We also found this to be the case for mutations affecting the GABA vesicular transporter-encoding gene unc-47 (Figure 4B), which is required for uptake of GABA into synaptic vesicles prior to neurotransmitter release. Mutations in unc-25 and unc-47 should reduce the release of GABA from GABAergic neurons (McIntire et al. 1993a). Interestingly, mutations in the GABAA receptor-encoding gene, unc-49, or in either of the GABAB metabotropic receptor subunits, gbb-1 and gbb-2, do not alter the sensitivity to ethanol for EHC (Figure 4, D–F). Finally, we examined the effect of mutations in the unc-30 gene, which alters the developmental fate of the type D GABAergic motorneurons, such that they no longer express unc-25 and unc-47 (Eastman et al. 1999). Unexpectedly, the unc-30 mutants showed a wild-type response to EHC (Figure 4C). We tested the possibility that the unc-25 and unc-47 mutants were already hypercontracted due to the lack of inhibition caused by a decrease in GABA signaling. In such a case, it may be that the GABA signaling mutants would not be able to contract further or to the same degree as WT animals, resulting in a floor effect in our measurements of EHC. To test this, we exposed unc-25, unc-47, and unc-49 mutant animals to aldicarb (0.5 or 1.0 mM), an acetylcholinesterase inhibitor that causes progressive hypercontraction and paralysis of C. elegans (Mahoney et al. 2006). All of the mutants, when exposed to aldicarb, contracted to a greater degree than they did in the presence of ethanol (Figure S2), confirming that our measurements were not confounded by a floor effect. The resistance to EHC shown by the unc-25 and unc-47 mutants suggests a role for GABA in the action of ethanol that generates hypercontraction, while the lack of effect of the unc-30 mutants suggests a role outside of the type D neurons.
Figure 4.
Loss of UNC-25 or UNC-47 function results in resistance to ethanol-induced hypercontraction. (A) GABA synthesis-defective unc-25 mutants displayed a significant reduction in the hypercontraction phenotype compared to wild-type animals (e156, ***P < 0.001; sa94, ^^P < 0.01, ^^^P < 0.001; n2569, #P < 0.05, ###P < 0.001; n = 6). (B) Loss of function of the vesicular GABA transporter, UNC-47, resulted in a significant decrease in the EHC phenotype (e307, *P < 0.05; n = 6; gk192, ^P < 0.05, ^^^P < 0.001; n = 6). (C) Mutations affecting unc-30, which encodes a transcription factor responsible for development of type D GABAergic motor neurons, did not alter EHC (P > 0.05; n = 6). (D–F). Mutants with a loss of signaling through GABAA (unc-49) or GABAB (gbb-1, gbb-2) were not significantly different from wild-type animals for EHC (P > 0.05; n = 5).
Neuropeptide signaling is not required for hypercontraction
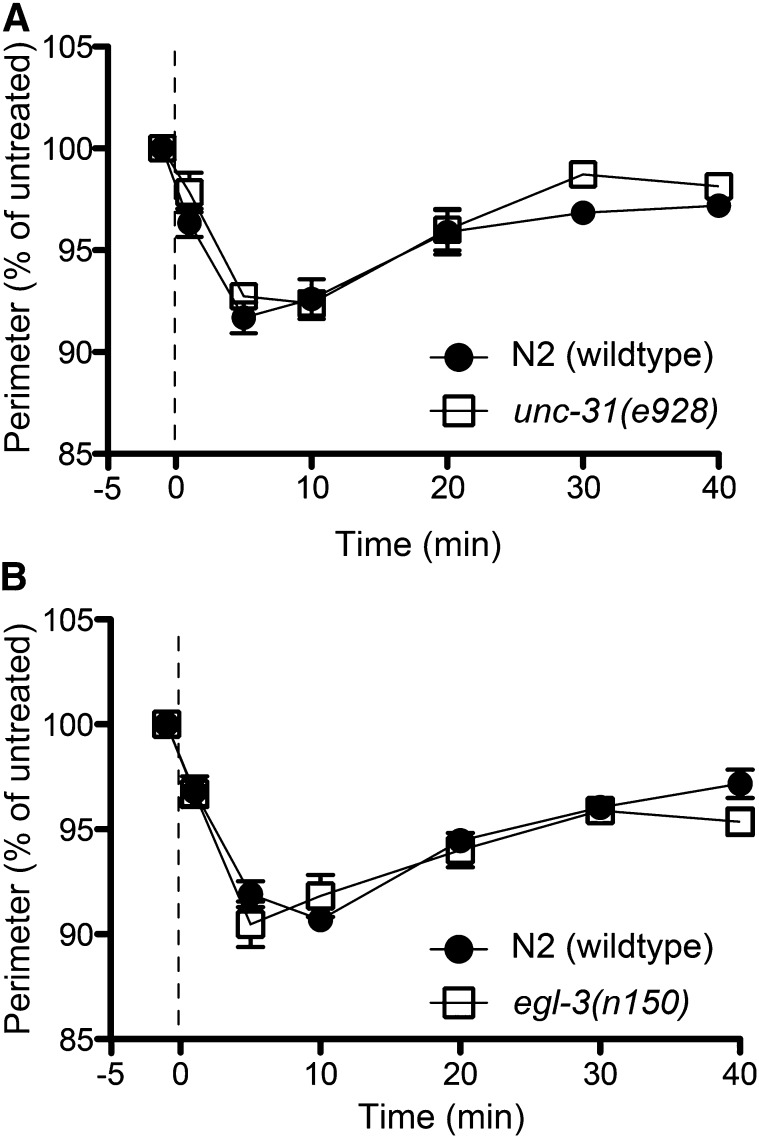
It has been reported that aldicarb treatment increases the release of the neuropeptide NLP-12 from the DVA neuron, which then potentiates cholinergic signaling at the NMJ of C. elegans (Hu et al. 2011). The DVA neuron is a stretch-sensitive sensory interneuron that requires the TRP-4 ion channel for its function. Worms exposed to ethanol exhibit a dose-dependent flattening of body bends (See Figure 1B) (Davies et al. 2003). We hypothesized that the EHC phenotype might be due to increased cholinergic signaling via NLP-12 release that is the result of the ethanol-induced reduction in body bend amplitude. We tested this hypothesis, using worms that are deficient in neuropeptide release and defective in DVA neuron function. Approximately 250 neuropeptides are released from dense core vesicles at the synaptic cleft in C. elegans (Li and Kim 2008), a process that requires UNC-31 and EGL-3. The unc-31 gene encodes the calcium-dependent activator protein for secretion (CAPS); loss of this protein inhibits the calcium-mediated fusion of dense core vesicles to the membrane, which prevents peptide release (Speese et al. 2007). egl-3 encodes a proprotein convertase type 2, which is required for the conversion of propeptides into active peptides (Kass et al. 2001). To test the role of neuropeptides in EHC, loss-of-function mutants for unc-31 and egl-3 were examined for sensitivity to EHC. Neither of these mutants exhibited a significantly different EHC phenotype compared with wild-type animals (Figure 5, A and B). To examine directly the role of the stretch pathway in EHC, we tested loss-of-function mutants of nlp-12 and trp-4 (Figure S3, A and B), which eliminate the NLP-12 response (Hu et al. 2011). Both of these mutants were not different from wild-type animals for the EHC response, indicating that the hypercontraction phenotype is not the result of increased cholinergic transmission through DVA activation. Taken in conjunction with the unc-31 and egl-3 data, this shows that neuropeptide signaling is unlikely to be involved in EHC.
Figure 5.
Neuropeptide signaling is not involved in ethanol-induced hypercontraction. (A and B) Mutations affecting two genes involved in neuropeptide release and activation, unc-31 and egl-3, respectively, exhibited similar contraction phenotypes to N2 (wild type) to 400 mM ethanol (P > 0.05; n = 6).
eat-6(eg200) is defective in the development of acute functional tolerance to EHC
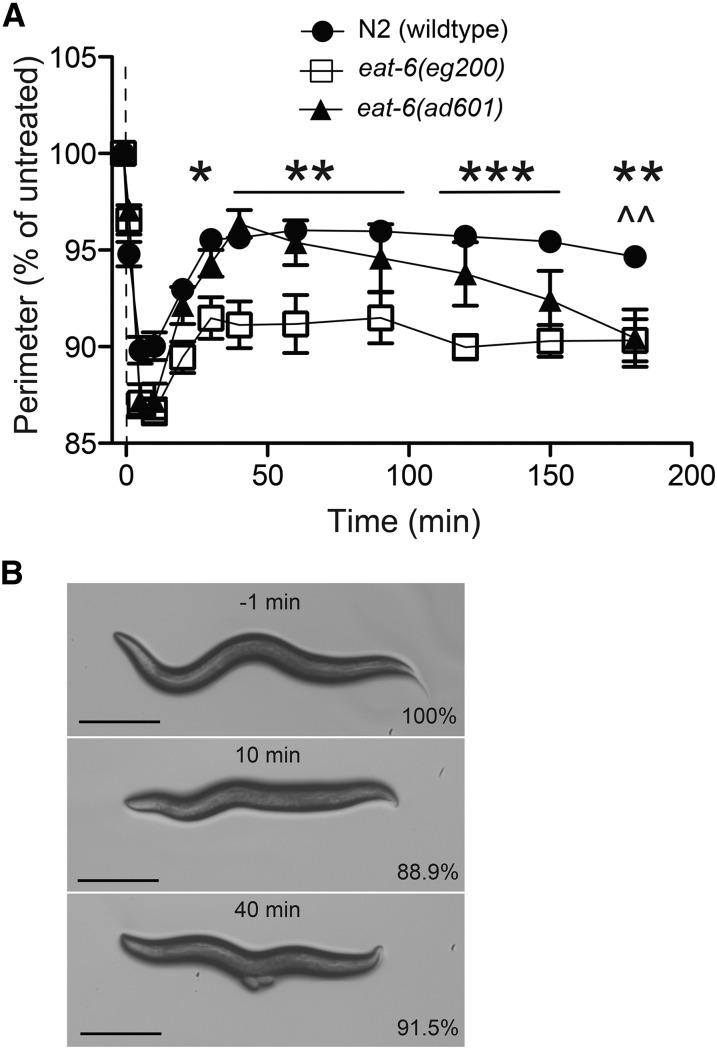
In a forward genetic screen, using ethyl methanesulfonate mutagenesis to generate random mutations, we isolated mutants that are hypersensitive to the effects of ethanol on the speed of locomotion. One of these mutants, eg200, was isolated on the basis of having significantly slower speeds on ethanol than wild-type siblings (Figure 6, A and B). However, the most striking aspect of the response to ethanol of the eg200 mutant animals was their continued hypercontraction for as long as they remained exposed to ethanol (Figure 7A), as opposed to the relatively rapid recovery demonstrated by wild-type animals (Figure 7A). We pursued identification of the gene affected by the eg200 mutation to better understand the second phase of EHC, the recovery (or acute functional tolerance) phase.
Figure 7.
Two eat-6 mutants show defects in the recovery from ethanol-induced hypercontraction. (A) N2 (wild type), eat-6(eg200), and eat-6(ad601) contract to a maximum degree in the first 10 min. N2 (wild type) and eat-6(ad601) recover while eat-6(eg200) remains significantly smaller after 40 min on 400 mM ethanol. eat-6(ad601) fails to maintain the recovery of contraction state; at 180 min both eat-6 alleles were significantly contracted compared to N2 (eg200, *P < 0.05, **P < 0.01, ***P < 0.001; ad601, ^^P < 0.01; n = 6). (B) Images of the same eat-6(eg200) worm treated with 400 mM ethanol at −1, 10, and 40 min. Percentages indicate percentage of the untreated perimeter at −1 min. Bar, 250 μm.
We used genetics to map the eg200 mutation to a region between lin-25 and the dimorphic SNP, snp_FF57B1[3] on chromosome V (Figure 6D) (see Materials and Methods). We examined the proposed functions, where known, of the 228 predicted genes in the ∼560-kb eg200-containing interval for candidate genes whose function might account for the hypersensitivity to and prolonged recovery from EHC. We investigated the shw-3 Shaw-class potassium channel as an eg200 candidate gene. Sequencing of the shw-3 gene amplified from eg200 genomic DNA did not reveal any mutations in the shw-3 coding sequence (data not shown). We next sequenced the eat-6 gene, which encodes the α-subunit of the Na+/K+-ATPase (Davis et al. 1995), which is an important regulator of excitable cell activity, and found a nucleotide substitution mutation that alters the codon for a highly conserved amino acid (glycine 77 in the EAT-6 protein) (Figure 6E), suggesting that the eg200 mutation affects the eat-6 gene.
The eg200 mutant animals do not have the starved appearance found with some other mutant alleles of the eat-6 gene (Avery 1993), but the eg200 animals are smaller and slower growing than wild-type animals, which is consistent with mutants that have feeding defects (Avery 1993). Some eat-6 mutants have been reported to be hypersensitive to the AChR agonist levamisole (Doi and Iwasaki 2008; Govorunova et al. 2010). We found that eg200 mutant animals are hypersensitive to the paralyzing effects of levamisole (Figure 6C), consistent with the eg200 mutant having a defect in the eat-6 gene. Although eg200 mutants do not have a starved appearance, they do have a reduction in pharyngeal pumping rate (Table S3), similar in severity to another eat-6 mutant, ad601, considered to be a weak allele for both starvation and pharyngeal pumping phenotypes (Davis et al. 1995; Govorunova et al. 2010). Two additional eat-6 alleles, ad467 and ad997, are reported to be stronger alleles with respect to mutant pharyngeal pumping and starvation phenotypes (Davis et al. 1995). We examined all three eat-6 alleles for altered recovery from EHC (Figure 7 and Figure S4B).
The eat-6(ad601) mutants contracted to the same extent as the eg200 mutant animals; however, unlike eg200 animals, the ad601 mutants initially recovered at the same rate as wild-type animals (Figure 7A). Interestingly, over the course of the next 2 hr of ethanol exposure, the ad601 mutant animals slowly contracted again to the same level as eg200 mutants, while wild-type animals did not undergo a second contraction over this time course (Figure 7A). We confirmed that eg200 is an allele of the eat-6 gene based on a failure to complement the ad601 for the EHC phenotype (Figure S4A). The trans-heterozygote eg200/ad601 displayed an EHC phenotype more similar to ad601 homozygotes; the eg200/ad601 animals initially recovered from the contracted state only to contract again at late time points of ethanol exposure.
Surprisingly, the two strong eat-6 mutants, ad467 and ad997, showed wild-type responses to EHC (Figure S4B). This result suggests that the strengths of the starvation and pharyngeal pumping mutant phenotypes are inversely related to the strength of the EHC phenotype. The same inverse relationship has been reported for sensitivity to the acetylcholinesterase inhibitor, aldicarb; ad601 has been reported to be more sensitive to the paralytic effects of aldicarb than the stronger alleles that cause visible starvation (Doi and Iwasaki 2008; Govorunova et al. 2010). We examined each of the eat-6 mutants for aldicarb sensitivity and found that eg200 was significantly more sensitive than any of the three remaining eat-6 alleles (Figure S4C). Under the conditions we used (1 mM aldicarb, no food), ad601 did not appear more sensitive to aldicarb than ad467 as has been reported (Doi and Iwasaki 2008; Govorunova et al. 2010), although in our assay all three mutations were more sensitive to aldicarb than wild-type animals (Figure S4C).
While the two mutants, eg200 and ad601, do not share identical EHC phenotypes, their ethanol responses are consistent with the EAT-6 Na+/K+-ATPase playing a role in the development and maintenance of tolerance to EHC.
EHC is distinct from previously identified ethanol responses
Mutations in multiple genes have previously been shown to generate altered phenotypes compared to wild-type animals upon treatment with ethanol (Davies et al. 2003, 2004; Kayser et al. 2003; Kapfhamer et al. 2008; Graham et al. 2009; Speca et al. 2010; Bettinger et al. 2012; Bhandari et al. 2012; Jee et al. 2013; Raabe et al. 2014). We examined two of these genes that have robust mutant ethanol response phenotypes, slo-1 and npr-1. Loss-of-function mutations in the BK potassium channel-encoding gene, slo-1, produce a significant level of resistance to the acute depressing effects of ethanol on locomotion speed and egg-laying frequency (Davies et al. 2003). We tested the null mutant slo-1(eg142) for its EHC response and found that slo-1(eg142) mutant animals have a wild-type level of EHC (Figure S3C). This suggests that the nAChR-mediated EHC effect is independent of the actions of ethanol that are mediated by SLO-1. Loss-of-function mutations in the neuropeptide Y receptor-like-encoding gene, npr-1, produce a more rapid development of acute functional tolerance to the depressing effects of ethanol on locomotion within an ethanol exposure session compared with wild-type animals (Davies et al. 2004). We tested whether the null mutant npr-1(ky13) would also alter the rate of development of tolerance to the EHC effect. As with the slo-1 mutant, the npr-1(ky13) mutant animals displayed wild-type sensitivity and rate of recovery to the EHC effect (Figure S3D), suggesting that the tolerance to the effects of ethanol on locomotion is independent of the tolerance that develops to EHC.
Discussion
The excitatory effects of ethanol have been phenotypically characterized in several different species, yet the molecular mechanism by which ethanol acts as an excitatory drug remains uncharacterized (Gingras and Cools 1996; Addicott et al. 2007; Arias et al. 2009; Beckstead and Phillips 2009; Kong et al. 2010; Rose et al. 2013). Using the model organism C. elegans, we developed a hypercontraction assay to study both the excitatory effects of ethanol and the tolerance that develops to these effects. This assay describes an effect of ethanol that is distinct from previously identified ethanol effects in the worm. Using an in vivo genetic approach, we were able to identify cholinergic and GABAergic signaling as being involved in mediating this excitatory effect. We identified an unusual mutation in the eat-6 Na+/K+-ATPase-encoding gene that prevented the development of acute functional tolerance to this effect.
Cholinergic signaling
Our data support a model in which exogenous administration of ethanol augments cholinergic signaling, causing a decrease in body length, presumably via an increase in the acetylcholine-mediated contraction of the body wall muscle. The nonselective and noncompetitive nAChR antagonist mecamylamine significantly blocked the EHC phenotype (Figure 2A), identifying a requirement for normal AChR function in this effect. Furthermore, animals carrying loss-of-function mutations in the genes encoding choline acetyltransferase cha-1 and the vesicular acetylcholine transporter unc-17, which result in low levels of ACh release into the synaptic cleft (Rand 2007), showed significant resistance to the EHC effect (Figure 2, B and C). These data support the hypothesis that ethanol increases cholinergic signaling to bring about EHC and that the availability of ACh for synaptic release is important for the degree of EHC.
We favored a hypothesis in which ethanol acts through specific AChRs because previous research has linked specific mammalian AChR subunits with electrophysiological and behavioral effects of ethanol. In vitro studies have shown direct effects of ethanol on nAChR currents, including potentiating effects (Cardoso et al. 1999; Godden et al. 2001). Our data indicate that there is a requirement for wild-type ACh availability for EHC and are consistent with the hypothesis that ethanol potentiates normal ACh signaling to generate EHC. Consistent with our mecamylamine result, it has been shown that mecamylamine can inhibit the locomotor-stimulation effects of ethanol in mice (Kamens and Phillips 2008). Our data indicate that unc-63, which encodes a nAChR α-subunit, is necessary for wild-type levels of EHC (Figure 3A). A mutation in unc-38, a paralog of unc-63, trended toward resistance of the EHC phenotype (Figure 3B), indicating that these related α-subunits may both play a role in EHC. The genes identified as orthologs of unc-63 and unc-38 in the Homo sapiens genome are CHRNA2 (α2), CHRNA3 (α3), CHRNA4 (α4), and CHRNA6 (α6) (Shaye and Greenwald 2011). Electrophysiological activities of specific subunits (α2β2, α2β4, α4β2, and α4β4) are enhanced in the presence of ACh and ethanol when measured in Xenopus oocytes (Cardoso et al. 1999; Borghese et al. 2003). The use of receptor-specific antagonists suggests that α3β2- or β3-containing receptors are involved in ethanol-induced locomotor stimulation in mice (Jerlhag et al. 2006), whereas α4β2- and α6-containing receptors are not (Larsson et al. 2002; Jerlhag et al. 2006). The involvement of α3 in locomotor stimulation agrees with genetic mapping and expression studies that suggest a role for the nAChR α3-subunit-encoding gene CHRNA3 in ethanol-induced locomotor stimulation in mice (Kamens et al. 2009). Behaviorally, heterozygotes containing an α3-knockout mutation were more sensitive to the depressant effects of ethanol than wild-type mice (Kamens et al. 2009), which suggests a possible loss of excitatory effects. Human genetics studies have found association with a cluster of three AChR-encoding genes that includes the α3-encoding gene and symptoms of alcohol abuse/dependence and with an acute level of response to alcohol (Joslyn et al. 2008; Chen et al. 2009; Sherva et al. 2010). That UNC-63 is considered to be the C. elegans ortholog of α3 suggests that there may be significant conservation between the ethanol-induced stimulation effect in mice, alcohol abuse/dependence in humans, and the EHC effect in worms that we have described here. Therefore, our data provide further support for the mammalian studies implicating specific α-subunits of the nAChRs in ethanol-induced stimulatory effects and strongly support this C. elegans phenotype as a relevant model of ethanol-induced excitatory effects.
Our finding that other subunits required for the levamisole-sensitive nAChR (Fleming et al. 1997; Culetto et al. 2004) are not required for EHC suggests that UNC-63 is acting in a different receptor to mediate EHC. We tested the only additional receptor currently known to include the UNC-63 subunit, the neuronal ACR-2 receptor (Jospin et al. 2009; Petrash et al. 2013), and found that loss of that receptor, through loss of function in the acr-2 gene, did not alter sensitivity to EHC. These results suggest that UNC-63 is likely to act in an as yet unidentified receptor(s) to mediate EHC.
GABA signaling in EHC
GABA signaling acts to oppose cholinergic signaling in coordinating body muscle contractions in C. elegans. We reasoned that if ethanol is acting through AChRs to mediate increased body muscle contraction, then loss of GABA signaling should enhance the effect of ethanol. Instead, we found the opposite result; a reduction in GABA synthesis and vesicular packaging produced via mutations in unc-25 and unc-47, respectively, reduced the degree of contraction induced by ethanol. We confirmed that this result was not due to a preexisting level of hypercontraction in the unc-25 and unc-47 mutants that limited the degree to which they could contract further (Figure S2A). One hypothesis to explain this outcome is that reduced GABA production and release may lead to a compensatory response that downregulates cholinergic signaling, which may then attenuate the degree to which ethanol can act through AChRs. If this is true, then the compensation must be specific in nature because it has been shown previously (Vashlishan et al. 2008) that GABAergic mutants, including unc-25, are actually more sensitive to aldicarb-induced paralysis, suggesting that with a reduction in GABA signaling, the body wall muscle is more sensitive to excitation rather than less sensitive as a compensatory model might predict. This suggests that there may be a more specific reduction in levels or activity of the UNC-63-containing receptor(s) in the unc-25 and unc-47 mutant backgrounds; such a reduction might not affect aldicarb sensitivity for muscle paralysis but would alter ethanol sensitivity. Also of interest are the differential effects on EHC of reduced GABA production and release in the unc-25 and unc-47 mutants compared with loss of either the major GABAA receptor UNC-49 or the two GABAB receptors GBB-1 and GBB-2. These results suggest the possibility that loss of both receptor types is required to mimic the reduction of GABA synthesis or, alternatively, that the impact of GABA on EHC is mediated by a third receptor type. The lack of effect of the loss of UNC-49, the major inhibitory receptor on the body muscle, suggests that the site of action for ethanol in producing EHC may not be directly on body muscle; instead an action on upstream neurons would better explain our observed data. We also found that unc-30 mutants, which lack GABA synthesis and vesicular packaging in the type D neurons (McIntire et al. 1993a), show a wild-type EHC phenotype. This result implicates the remaining GABAergic neurons (AVL, DVB, RIS, and RMEs) that are affected by unc-25 and unc-47 mutations but are unaffected in the unc-30 mutant animals. Of those neurons, AVL, DVB, and the four RMEs are primarily involved in defecation and in head foraging movement (McIntire et al. 1993b), and a GABAergic function associated with the RIS interneuron is yet to be identified. The mechanism by which decreased GABA synthesis and release reduces sensitivity to EHC remains to be identified.
eat-6 mutations affect tolerance to EHC
The eat-6(eg200) mutant demonstrates a prolonged response to EHC. The continued hypercontraction in the presence of ethanol by eat-6(eg200) and the return to hypercontraction following an initial recovery that is shown by the eat-6(ad601) mutant highlight the requirement for EAT-6 function in the acute functional tolerance that is occurring in wild-type animals to this effect of ethanol during the period of ethanol exposure.
The eat-6 gene encodes the α-subunit of the Na+/K+-ATPase (Davis et al. 1995), which pumps three sodium ions out of and two potassium ions into the cell. As a result, maintenance of a negative membrane potential is achieved as dictated by the Goldman equation (Hille 2001). Loss of function of this protein would be predicted to result in a depolarized and hyperexcitable cell (Davis et al. 1995). One possibility to explain the prolonged EHC phenotype in the eat-6(eg200) mutant animals is that eg200 is a loss-of-function allele that leads to a hyperexcitable state in cells activated by ethanol and, in combination with potentiation of cholinergic signaling by ethanol, the affected cells cannot compensate for the strong hyperactivation to achieve a homeostatic recovery. The strong levamisole and aldicarb hypersensitivity phenotypes associated with eg200 support the model that eg200 mutants are particularly sensitive to cholinergic activation. However, the idea of eg200 as a straightforward loss-of-function mutation is not supported by the EHC or aldicarb phenotypes displayed by other alleles of eat-6, some of which have stronger effects on pharyngeal pumping (Davis et al. 1995).
eat-6 mutants have been described as having several phenotypes, including defects in pharyngeal pumping, and altered sensitivities to aldicarb, levamisole, nicotine, and serotonin (Avery 1993; Davis et al. 1995; Doi and Iwasaki 2008; Govorunova et al. 2010). Different alleles of eat-6 have different constellations of phenotypes, suggesting that they do not fall into a simple allelic series representing an increasing degree of loss of function; instead different alleles have different strengths of effect on a variety of different neuromuscular phenotypes, to which we have added the development of tolerance to EHC (Table S3). The eg200 and ad601 mutations are both weak alleles for effects on starvation, and both produce hypersensitivity to levamisole, but they differ in the time course for their effect on the development of tolerance to EHC. The ad467 and ad997 mutations have strong effects on pharyngeal pumping and feeding, but have no detectable effect on EHC. All alleles show hypersensitivity to aldicarb-induced paralysis in our assay but eg200 shows the greatest level of hypersensitivity. These observations combine to suggest that the eat-6 mutation is unusual in comparison with other isolated eat-6 mutations.
An alternative model to explain the effect of the eg200 mutation is that tolerance to the EHC effect may require regulation of Na+/K+-ATPase function, and the eg200 mutation specifically affects the ability of the Na+/K+-ATPase to make such changes. The eg200 mutation is predicted to result in glycine to glutamic acid substitution at residue 77. The role of the equivalent conserved amino acid, Gly94, in the rat Na+/K+-ATPase has been investigated (Einholm et al. 2005). Substitution of Gly94 by Ala94 in the M1 transmembrane region interferes with binding of both Na+ and K+ by the enzyme. The results were particularly striking for the E2P conformational state of the enzyme, which had a ninefold reduced affinity for the K+ cation, resulting in a dysfunctional transporter (Einholm et al. 2005). The importance of that specific amino acid in enzyme function, particularly in certain conformations, may be significant in the development of tolerance to EHC.
One other possibility for the role of EAT-6 in altering cholinergic signaling and the sensitivity to, or recovery from, EHC is that EAT-6 has been found to act in an allele-specific and pump-independent manner to regulate nAChR localization at the postsynaptic membrane of the NMJ (Doi and Iwasaki 2008). The ad601 mutation was found to have the strongest effect on nAChR localization (Doi and Iwasaki 2008); we found that when exposed to ethanol, ad601 had a delayed return to a hypercontracted state following an initial wild-type-like recovery. The possibility that the number of UNC-63-containing receptors could be increased postsynaptically in eat-6(eg200) mutant animals might also account for a prolonged EHC effect, particularly if the regulation of the localization of those receptors was abnormal. The observation of increased levamisole and aldicarb sensitivity in the eat-6(eg200) mutant would also be consistent with eg200 acting in a similar manner to ad601 for its effects on the levamisole-sensitive receptor at the NMJ. The impact of eat-6(eg200) on the unknown ethanol-sensitive receptor(s) containing UNC-63 is worthy of future examination.
Conclusion
We identified an excitatory effect of ethanol that we have termed EHC in the model organism C. elegans and developed a novel software-assisted assay method to aid in data analysis. We used both genetic and pharmacological approaches to determine the molecular mechanisms involved in this ethanol-induced effect. With this in vivo model we found that the excitatory phenotype is cholinergic and functions through the UNC-63 α-nAChR subunit, which is orthologous to the mammalian α2, α3, α4, and α6 nAChR subunits. This excitation effect of ethanol characterizes a property of an unidentified receptor type because known receptors that include the UNC-63 subunit were not required for this effect. Furthermore, we show that levels of GABA release alter the degree of EHC; however, the mechanism involved does not appear to involve the well-characterized C. elegans GABAA or GABAB receptors. In addition to the acute effect of ethanol on excitation, we show that acute functional tolerance develops rapidly to the effect. We identify a missense mutation of eat-6 that alters a conserved amino acid in the Na+/K+-ATPase as having the effect of eliminating the development of tolerance to the ethanol effect, causing a prolonged hypercontracted phenotype. This suggests that the Na+/K+-ATPase plays a role in the development of tolerance to this ethanol effect. Genes previously shown to affect acute sensitivity and the development of acute functional tolerance to the depressive effect of ethanol on locomotion speed, slo-1 and npr-1, respectively, are not involved in the excitatory mechanism. Our data indicate that the mechanisms of ethanol-induced excitation and depression and the mechanisms of tolerance to these effects are different from each other.
Acknowledgments
The authors thank Steven L. McIntire, in whose laboratory this work was initiated. We thank Laura Mathies for helpful discussions. We thank S. Mitani (National BioResource Project) and the Caenorhabditis Genetics Center (CGC) for providing strains. The CGC is funded by National Institutes of Health (NIH) Office of Research Infrastructure Programs (OD010440). Funding for this work was provided by NIH grants AA016842 (to A.G.D. and J.C.B.), AA017070 (to J.C.B. and A.G.D.), UL1TR000058 (to A.G.D.), and DA007027 (to E.G.H.) and by ABMRF/The Foundation for Alcohol Research (A.G.D.).
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.171884/-/DC1.
Communicating editor: D. I. Greenstein
Literature Cited
- Addicott M. A., Marsh-Richard D. M., Mathias C. W., Dougherty D. M., 2007. The biphasic effects of alcohol: comparisons of subjective and objective measures of stimulation, sedation, and physical activity. Alcohol. Clin. Exp. Res. 31: 1883–1890. [DOI] [PubMed] [Google Scholar]
- Alaimo J. T., Davis S. J., Song S. S., Burnette C. R., Grotewiel M., et al. , 2012. Ethanol metabolism and osmolarity modify behavioral responses to ethanol in C. elegans. Alcohol. Clin. Exp. Res. 36: 1840–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfonso A., Grundahl K., Duerr J. S., Han H. P., Rand J. B., 1993. The Caenorhabditis elegans unc-17 gene: a putative vesicular acetylcholine transporter. Science 261: 617–619. [DOI] [PubMed] [Google Scholar]
- Arias C., Mlewski E. C., Molina J. C., Spear N. E., 2009. Ethanol induces locomotor activating effects in preweanling Sprague-Dawley rats. Alcohol 43: 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artal-Sanz M., de Jong L., and N. Tavernarakis, 2006. Caenorhabditis elegans: a versatile platform for drug discovery. Biotechnol. J. 1: 1405–1418. [DOI] [PubMed] [Google Scholar]
- Avery L., 1993. The genetics of feeding in Caenorhabditis elegans. Genetics 133: 897–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates R. G., Paabo M., Robinson R. A., 1963. Interpretation of pH measurements in alcohol-water solvents. J. Phys. Chem. 67: 1833–1838. [Google Scholar]
- Beckstead M. J., Phillips T. J., 2009. Mice selectively bred for high- or low-alcohol-induced locomotion exhibit differences in dopamine neuron function. J. Pharmacol. Exp. Ther. 329: 342–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettinger J. C., Leung K., Bolling M. H., Goldsmith A. D., Davies A. G., 2012. Lipid environment modulates the development of acute tolerance to ethanol in Caenorhabditis elegans. PLoS ONE 7: e35192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari P., Hill J. S., Farris S. P., Costin B., Martin I., et al. , 2012. Chloride intracellular channels modulate acute ethanol behaviors in Drosophila, Caenorhabditis elegans and mice. Genes Brain Behav. 11: 387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghese C. M., Henderson L. A., Bleck V., Trudell J. R., Harris R. A., 2003. Sites of excitatory and inhibitory actions of alcohols on neuronal alpha2beta4 nicotinic acetylcholine receptors. J. Pharmacol. Exp. Ther. 307: 42–52. [DOI] [PubMed] [Google Scholar]
- Brown L. A., Jones A. K., Buckingham S. D., Mee C. J., Sattelle D. B., 2006. Contributions from Caenorhabditis elegans functional genetics to antiparasitic drug target identification and validation: nicotinic acetylcholine receptors, a case study. Int. J. Parasitol. 36: 617–624. [DOI] [PubMed] [Google Scholar]
- Cardoso R. A., Brozowski S. J., Chavez-Noriega L. E., Harpold M., Valenzuela C. F., et al. , 1999. Effects of ethanol on recombinant human neuronal nicotinic acetylcholine receptors expressed in Xenopus oocytes. J. Pharmacol. Exp. Ther. 289: 774–780. [PubMed] [Google Scholar]
- Chen X., Chen J., Williamson V. S., An S. S., Hettema J. M., et al. , 2009. Variants in nicotinic acetylcholine receptors alpha5 and alpha3 increase risks to nicotine dependence. Am. J. Med. Genet. B Neuropsychiatr. Genet. 150B: 926–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe K. P., Strange K., 2007. Evolutionarily conserved WNK and Ste20 kinases are essential for acute volume recovery and survival after hypertonic shrinkage in Caenorhabditis elegans. Am. J. Physiol. Cell Physiol. 293: C915–C927. [DOI] [PubMed] [Google Scholar]
- Culetto E., Baylis H. A., Richmond J. E., Jones A. K., Fleming J. T., et al. , 2004. The Caenorhabditis elegans unc-63 gene encodes a levamisole-sensitive nicotinic acetylcholine receptor alpha subunit. J. Biol. Chem. 279: 42476–42483. [DOI] [PubMed] [Google Scholar]
- Davies A. G., Pierce-Shimomura J. T., Kim H., Vanhoven M. K., Thiele T. R., et al. , 2003. A central role of the BK potassium channel in behavioral responses to ethanol in C. elegans. Cell 115: 655–666. [DOI] [PubMed] [Google Scholar]
- Davies A. G., Bettinger J. C., Thiele T. R., Judy M. E., McIntire S. L., 2004. Natural variation in the npr-1 gene modifies ethanol responses of wild strains of C. elegans. Neuron 42: 731–743. [DOI] [PubMed] [Google Scholar]
- Davies A. G., Friedberg R. I., Gupta H., Chan C.-L., Shelton K. L., et al. , 2012. Different genes influence toluene- and ethanol-induced locomotor impairment in C. elegans. Drug Alcohol Depend. 122: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. W., Somerville D., Lee R. Y., Lockery S., Avery L., et al. , 1995. Mutations in the Caenorhabditis elegans Na,K-ATPase alpha-subunit gene, eat-6, disrupt excitable cell function. J. Neurosci. 15: 8408–8418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. W., Hammarlund M., Harrach T., Hullett P., Olsen S., et al. , 2005. Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics 6: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M., Iwasaki K., 2008. Na+/K+ ATPase regulates the expression and localization of acetylcholine receptors in a pump activity-independent manner. Mol. Cell. Neurosci. 38: 548–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman C., Horvitz H. R., Jin Y., 1999. Coordinated transcriptional regulation of the unc-25 glutamic acid decarboxylase and the unc-47 GABA vesicular transporter by the Caenorhabditis elegans UNC-30 homeodomain protein. J. Neurosci. 19: 6225–6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einholm A. P., Toustrup-Jensen M., Andersen J. P., Vilsen B., 2005. Mutation of Gly-94 in transmembrane segment M1 of Na+,K+-ATPase interferes with Na+ and K+ binding in E2P conformation. Proc. Natl. Acad. Sci. USA 102: 11254–11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming J. T., Squire M. D., Barnes T. M., Tornoe C., Matsuda K., et al. , 1997. Caenorhabditis elegans levamisole resistance genes lev-1, unc-29, and unc-38 encode functional nicotinic acetylcholine receptor subunits. J. Neurosci. 17: 5843–5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras M. A., Cools A. R., 1996. Analysis of the biphasic locomotor response to ethanol in high and low responders to novelty: a study in Nijmegen Wistar rats. Psychopharmacology 125: 258–264. [DOI] [PubMed] [Google Scholar]
- Godden E. L., Harris R. A., Dunwiddie T. V., 2001. Correlation between molecular volume and effects of n-alcohols on human neuronal nicotinic acetylcholine receptors expressed in Xenopus oocytes. J. Pharmacol. Exp. Ther. 296: 716–722. [PubMed] [Google Scholar]
- Govorunova E. G., Moussaif M., Kullyev A., Nguyen K. C., Mcdonald T. V., et al. , 2010. A homolog of FHM2 is involved in modulation of excitatory neurotransmission by serotonin in C. elegans. PLoS ONE 5: e10368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham M. E., Edwards M. R., Holden-Dye L., Morgan A., Burgoyne R. D., et al. , 2009. UNC-18 modulates ethanol sensitivity in Caenorhabditis elegans. Mol. Biol. Cell 20: 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. A., Trudell J. R., Mihic S. J., 2008. Ethanol’s molecular targets. Sci. Signal. 1: re7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath A., Madden P., Bucholz K., Dinwiddie S., Slutske W., et al. , 1999. Genetic differences in alcohol sensitivity and the inheritance of alcoholism risk. Psychol. Med. 29: 1069–1081. [DOI] [PubMed] [Google Scholar]
- Hille B., 2001. Ion Channels of Excitable Membranes. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Holstein S. E., Dobbs L., Phillips T. J., 2009. Attenuation of the stimulant response to ethanol is associated with enhanced ataxia for a GABAA, but not a GABAB, receptor agonist. Alcohol. Clin. Exp. Res. 33: 108–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu D. R., Meyer B. J., 1994. The dpy-30 gene encodes an essential component of the Caenorhabditis elegans dosage compensation machinery. Genetics 137: 999–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Pym E. C., Babu K., Vashlishan Murray A. B., Kaplan J. M., 2011. A neuropeptide-mediated stretch response links muscle contraction to changes in neurotransmitter release. Neuron 71: 92–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Cheng H. J., Tessier-Lavigne M., Jin Y., 2002. MAX-1, a novel PH/MyTH4/FERM domain cytoplasmic protein implicated in netrin-mediated axon repulsion. Neuron 34: 563–576. [DOI] [PubMed] [Google Scholar]
- Jee C., Lee J., Lim J. P., Parry D., Messing R. O., et al. , 2013. SEB-3, a CRF receptor-like GPCR, regulates locomotor activity states, stress responses and ethanol tolerance in Caenorhabditis elegans. Genes Brain Behav. 12: 250–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerlhag E., Grotli M., Luthman K., Svensson L., Engel J. A., 2006. Role of the subunit composition of central nicotinic acetylcholine receptors for the stimulatory and dopamine-enhancing effects of ethanol. Alcohol Alcohol. 41: 486–493. [DOI] [PubMed] [Google Scholar]
- Jones A. K., Davis P., Hodgkin J., Sattelle D. B., 2007. The nicotinic acetylcholine receptor gene family of the nematode Caenorhabditis elegans: an update on nomenclature. Invert. Neurosci. 7: 129–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joslyn G., Brush G., Robertson M., Smith T. L., Kalmijn J., et al. , 2008. Chromosome 15q25.1 genetic markers associated with level of response to alcohol in humans. Proc. Natl. Acad. Sci. USA 105: 20368–20373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jospin M., Qi Y. B., Stawicki T. M., Boulin T., Schuske K. R., et al. , 2009. A neuronal acetylcholine receptor regulates the balance of muscle excitation and inhibition in Caenorhabditis elegans. PLoS Biol. 7: e1000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletta T., Hengartner M. O., 2006. Finding function in novel targets: C. elegans as a model organism. Nat. Rev. Drug Discov. 5: 387–398. [DOI] [PubMed] [Google Scholar]
- Kalu N., Ramchandani V. M., V. Marshall, D. Scott, C. Ferguson et al, 2012. Heritability of level of response and association with recent drinking history in nonalcohol-dependent drinkers. Alcohol. Clin. Exp. Res. 36: 522–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamens H. M., Phillips T. J., 2008. A role for neuronal nicotinic acetylcholine receptors in ethanol-induced stimulation, but not cocaine- or methamphetamine-induced stimulation. Psychopharmacology 196: 377–387. [DOI] [PubMed] [Google Scholar]
- Kamens H. M., Mckinnon C. S., Li N., Helms M. L., Belknap J. K., et al. , 2009. The alpha 3 subunit gene of the nicotinic acetylcholine receptor is a candidate gene for ethanol stimulation. Genes Brain Behav. 8: 600–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapfhamer D., Bettinger J. C., Davies A. G., Eastman C. L., Smail E. A., et al. , 2008. Loss of RAB-3/A in Caenorhabditis elegans and the mouse affects behavioral response to ethanol. Genes Brain Behav. 7: 669–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass J., Jacob T. C., Kim P., Kaplan J. M., 2001. The EGL-3 proprotein convertase regulates mechanosensory responses of Caenorhabditis elegans. J. Neurosci. 21: 9265–9272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser E. B., Hoppel C. L., Morgan P. G., Sedensky M. M., 2003. A mutation in mitochondrial complex I increases ethanol sensitivity in Caenorhabditis elegans. Alcohol. Clin. Exp. Res. 27: 584–592. [DOI] [PubMed] [Google Scholar]
- Khattab I. S., Bandarkar F., Fakhree M. A. A., Jouyban A., 2012. Density, viscosity, and surface tension of water+ethanol mixtures from 293 to 323K. Korean J. Chem. Eng. 29: 812–817. [Google Scholar]
- Kong E. C., Woo K., Li H., Lebestky T., Mayer N., et al. , 2010. A pair of dopamine neurons target the D1-like dopamine receptor DopR in the central complex to promote ethanol-stimulated locomotion in Drosophila. PLoS ONE 5: e9954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse L. C., Linsenbardt D. N., Boehm S. L., 2nd, 2012. Positive allosteric modulation of the GABAB receptor by GS39783 attenuates the locomotor stimulant actions of ethanol and potentiates the induction of locomotor sensitization. Alcohol 46: 455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok T. C., Ricker N., Fraser R., Chan A. W., Burns A., et al. , 2006. A small-molecule screen in C. elegans yields a new calcium channel antagonist. Nature 441: 91–95. [DOI] [PubMed] [Google Scholar]
- Larsson A., Svensson L., Soderpalm B., Engel J. A., 2002. Role of different nicotinic acetylcholine receptors in mediating behavioral and neurochemical effects of ethanol in mice. Alcohol 28: 157–167. [DOI] [PubMed] [Google Scholar]
- Lewis J. A., Wu C. H., Berg H., Levine J. H., 1980. The genetics of levamisole resistance in the nematode Caenorhabditis elegans. Genetics 95: 905–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C., and K. Kim, 2008 Neuropeptides (April 7, 2008), WormBook, ed. The C. elegans Research Community, WormBook, 10.1895/wormbook.1.142.1, http://www.wormbook.org.
- Mahoney T. R., Luo S., Nonet M. L., 2006. Analysis of synaptic transmission in Caenorhabditis elegans using an aldicarb-sensitivity assay. Nat. Protoc. 1: 1772–1777. [DOI] [PubMed] [Google Scholar]
- McIntire S. L., Jorgensen E. M., Horvitz H. R., 1993a Genes required for GABA function in Caenorhabditis elegans. Nature 364: 334–337. [DOI] [PubMed] [Google Scholar]
- McIntire S. L., Jorgensen E. M., Kaplan J. M., Horvitz H. R., 1993b The GABAergic nervous system of Caenorhabditis elegans. Nature 364: 337–341. [DOI] [PubMed] [Google Scholar]
- Mitchell P., Mould R., Dillon J., Glautier S., Andrianakis I., et al. , 2010. A differential role for neuropeptides in acute and chronic adaptive responses to alcohol: behavioural and genetic analysis in Caenorhabditis elegans. PLoS ONE 5: e10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. H., Bull K., Glautier S., Hopper N. A., Holden-Dye L., et al. , 2007. The concentration-dependent effects of ethanol on Caenorhabditis elegans behaviour. Pharmacogenomics J. 7: 411–417. [DOI] [PubMed] [Google Scholar]
- Newlin D. B., Thomson J. B., 1990. Alcohol challenge with sons of alcoholics: a critical review and analysis. Psychol. Bull. 108: 383–402. [DOI] [PubMed] [Google Scholar]
- Nurrish S., Segalat L., Kaplan J. M., 1999. Serotonin inhibition of synaptic transmission: Galpha(0) decreases the abundance of UNC-13 at release sites. Neuron 24: 231–242. [DOI] [PubMed] [Google Scholar]
- Olsen R., Li G., Wallner M., Trudell J., Bertaccini E., et al. , 2014. Structural models of ligand-gated ion channels: sites of action for anesthetics and ethanol. Alcohol. Clin. Exp. Res. 38: 595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrash H. A., Philbrook A., Haburcak M., Barbagallo B., Francis M. M., 2013. ACR-12 ionotropic acetylcholine receptor complexes regulate inhibitory motor neuron activity in Caenorhabditis elegans. J. Neurosci. 33: 5524–5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips T. J., Shen E. H., 1996. Neurochemical bases of locomotion and ethanol stimulant effects. Int. Rev. Neurobiol. 39: 243–282. [DOI] [PubMed] [Google Scholar]
- Ponomarev I., Crabbe J. C., 2002. Ethanol-induced activation and rapid development of tolerance may have some underlying genes in common. Genes Brain Behav. 1: 82–87. [DOI] [PubMed] [Google Scholar]
- Raabe R., Mathies L., Davies A., Bettinger J., 2014. The omega-3 fatty acid eicosapentaenoic acid is required for normal alcohol response behaviors in C. elegans. PLoS ONE 9: e105999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand, J. B., 2007 Acetylcholine (January 30, 2007), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.131.1, http://www.wormbook.org.
- Rand J. B., Russell R. L., 1984. Choline acetyltransferase-deficient mutants of the nematode Caenorhabditis elegans. Genetics 106: 227–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond J. E., Jorgensen E. M., 1999. One GABA and two acetylcholine receptors function at the C. elegans neuromuscular junction. Nat. Neurosci. 2: 791–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose J. H., Calipari E. S., Mathews T. A., Jones S. R., 2013. Greater ethanol-induced locomotor activation in DBA/2J vs. C57BL/6J mice is not predicted by presynaptic striatal dopamine dynamics. PLoS ONE 8: e83852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit M. A., 1994. Low level of response to alcohol as a predictor of future alcoholism. Am. J. Psychiatry 151: 184–189. [DOI] [PubMed] [Google Scholar]
- Shaye D., Greenwald I., 2011. OrthoList: a compendium of C. elegans genes with human orthologs. PLoS ONE 6: e20085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherva R., Kranzler H. R., Yu Y., Logue M. W., Poling J., et al. , 2010. Variation in nicotinic acetylcholine receptor genes is associated with multiple substance dependence phenotypes. Neuropsychopharmacology 35: 1921–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderpalm B., Ericson M., Olausson P., Blomqvist O., Engel J. A., 2000. Nicotinic mechanisms involved in the dopamine activating and reinforcing properties of ethanol. Behav. Brain Res. 113: 85–96. [DOI] [PubMed] [Google Scholar]
- Spanagel R., 2009. Alcoholism: a systems approach from molecular physiology to addictive behavior. Physiol. Rev. 89: 649–705. [DOI] [PubMed] [Google Scholar]
- Speca D. J., Chihara D., Ashique A. M., Bowers M. S., Pierce-Shimomura J. T., et al. , 2010. Conserved role of unc-79 in ethanol responses in lightweight mutant mice. PLoS Genet. 6: e1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speese S., Petrie M., Schuske K., Ailion M., Ann K., et al. , 2007. UNC-31 (CAPS) is required for dense-core vesicle but not synaptic vesicle exocytosis in Caenorhabditis elegans. J. Neurosci. 27: 6150–6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan K. A., Curtis D. E., Mckusick K. B., Voinov A. V., Mapa F. A., et al. , 2002. High-throughput gene mapping in Caenorhabditis elegans. Genome Res. 12: 1100–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topper S., Aguilar S., Topper V., Elbel E., Pierce-Shimomura J. T., 2014. Alcohol disinhibition of behaviors in C. elegans. PLoS ONE 9: e92965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touroutine D., Fox R. M., Von Stetina S. E., A. Burdina, D. M. Miller, 3rd et al, 2005. acr-16 encodes an essential subunit of the levamisole-resistant nicotinic receptor at the Caenorhabditis elegans neuromuscular junction. J. Biol. Chem. 280: 27013–27021. [DOI] [PubMed] [Google Scholar]
- Towers P. R., Edwards B., Richmond J. E., Sattelle D. B., 2005. The Caenorhabditis elegans lev-8 gene encodes a novel type of nicotinic acetylcholine receptor alpha subunit. J. Neurochem. 93: 1–9. [DOI] [PubMed] [Google Scholar]
- Tuck S., Greenwald I., 1995. lin-25, a gene required for vulval induction in Caenorhabditis elegans. Genes Dev. 9: 341–357. [DOI] [PubMed] [Google Scholar]
- Van Swinderen B., Saifee O., Shebester L., Roberson R., Nonet M. L., et al. , 1999. A neomorphic syntaxin mutation blocks volatile-anesthetic action in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 96: 2479–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashlishan A. B., Madison J. M., Dybbs M., Bai J., Sieburth D., et al. , 2008. An RNAi screen identifies genes that regulate GABA synapses. Neuron 58: 346–361. [DOI] [PubMed] [Google Scholar]
- Volavka J., Czobor P., Goodwin D. W., Gabrielli W. F., Jr, Penick E. C., et al. , 1996. The electroencephalogram after alcohol administration in high-risk men and the development of alcohol use disorders 10 years later. Arch. Gen. Psychiatry 53: 258–263. [DOI] [PubMed] [Google Scholar]
- Wicks S. R., Yeh R. T., Gish W. R., Waterston R. H., Plasterk R. H., 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28: 160–164. [DOI] [PubMed] [Google Scholar]