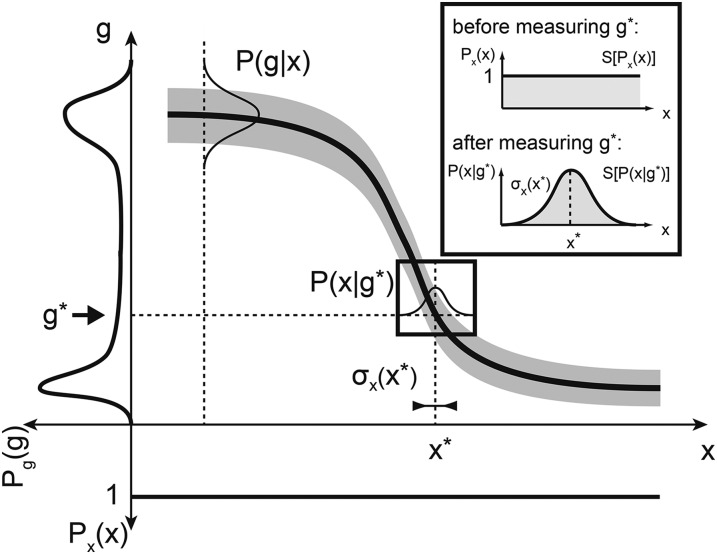
Figure 3.
The mathematics of positional information and positional error for one gene. The mean profile of gene g (thick solid line) and its variability (shaded envelope) across embryos are shown. Nuclei are distributed uniformly along the AP axis; i.e., the prior distribution of nuclear positions Px(x) is uniform (shown at the bottom). The total distribution of expression levels across the embryo, Pg(g), is determined by averaging P(g|x) over all positions and is shown on the left. The positional information I(g; x) is related to these two distributions as in Equation 8. Inset shows decoding (i.e., estimating) the position of a nucleus from a measured expression level g*. Prior to the “measurement” all positions are equally likely. After observing the value g*, the positions consistent with this measured values are drawn from P(x|g*). The best estimate of the true position, x*, is at the peak of this distribution, and the positional error, σx(x), is the distribution’s width. Positional information I(g; x) can equally be computed from Px(x) and P(x|g) by Equation 10.