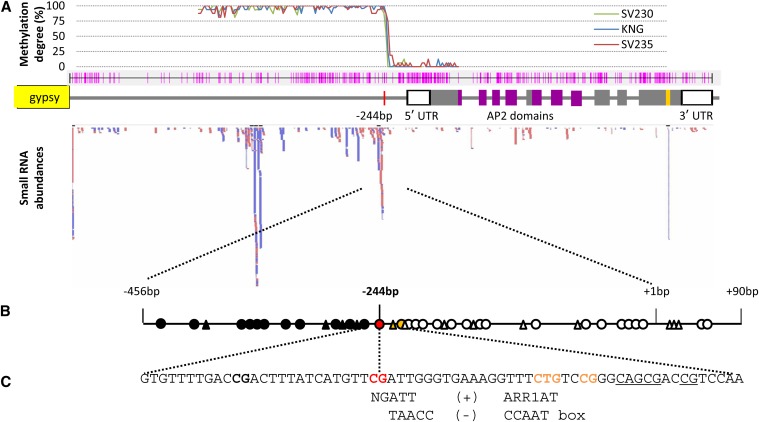
Figure 5.
Putative regulatory factors for cly1. (A) Global DNA methylation degree related to siRNA frequency. Methylation levels as revealed by the proportion of CpG sites methylated; the pink lines indicate the positions of CpG and CpNpG sites in the upstream and coding regions of cly1. The red bars indicate siRNA reads homologous to the cly1 genomic sequence, and the blue bars show complementary reads. (B) DNA methylation in the upstream region of cly1. Solid circles indicate hypermethylated CpG sites, the red circle identifies the CpG site correlated with cly1 transcription, and open circles show demethylated CpG sites; solid triangles indicate methylated CpNpG sites and open ones nonmethylated CpNpG sites. (C) Putative cis elements. NGATT: an ARR1 binding element (+ strand). TAACC: a CCAAT box (− strand).