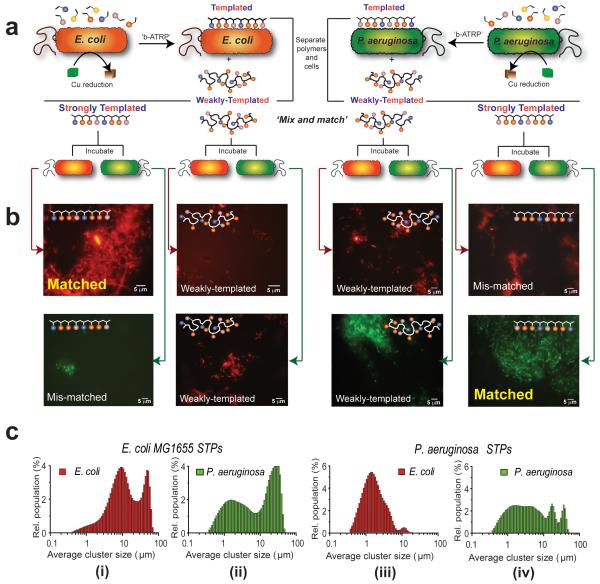
Figure 3. Demonstration of self-selective microbial binding by ‘bacteria-instructed polymers’.
In (A) the experimental design for the cell-selective polymer binding assays is shown. Bacteria (E. coli, depicted in orange, or P. aeruginosa, shown in green) were used to template bacterial-mediated synthesis to generate two polymer fractions, shown schematically below the cells. ‘Weakly-Templated Polymers’ (WTP) were obtained from the supernatant following cell centrifugation while the more tightly-bound ‘Strongly-Templated Polymers’ (STP), were recovered from the bacteria by a salt solution wash. In (B) are shown fluorescence micrographs of experiments where polymers templated with E coli MG1655 (expressing mCherry, orange-red) and P aeruginosa PAO1 (expressing GFP, green) were incubated with their ‘matched’ bacteria (i,e. fresh suspension of the cell strains used to template synthesis) and with ‘mismatched’ bacteria (i.e. P. aeruginosa suspensions added to polymers grown from E. coli and vice versa). Micrographs labelled ‘Control’ refer to incubations of cells with polymers grown in the absence of cells. The ‘mix and match’ combinations of bacteria, the polymers templated from them and the non-templated polymers are shown by connecting red arrows for E coli cells and green arrows for P. aeruginosa (images A to B). Cell aggregation by polymers, as shown in (B), is quantified in (C), with each graph in (C i-iv) relating to microscopy images of the ‘matched’ and ‘mis-matched’ combinations of bacteria and polymers shown in (B) above. The distributions of E. coli aggregates (C i. ‘matched’) are shifted to higher sizes with E. coli-templated polymers compared to P. aeruginosa clusters with the same polymer (C ii, ‘mis-matched’), while (C iv, ‘matched’) indicates P. aeruginosa-templated polymers induce greatest cluster sizes with P. aeruginosa compared to E. coli (C iii, ‘mis-matched’), confirming the enhanced binding of ‘bacteria-instructed polymers’.