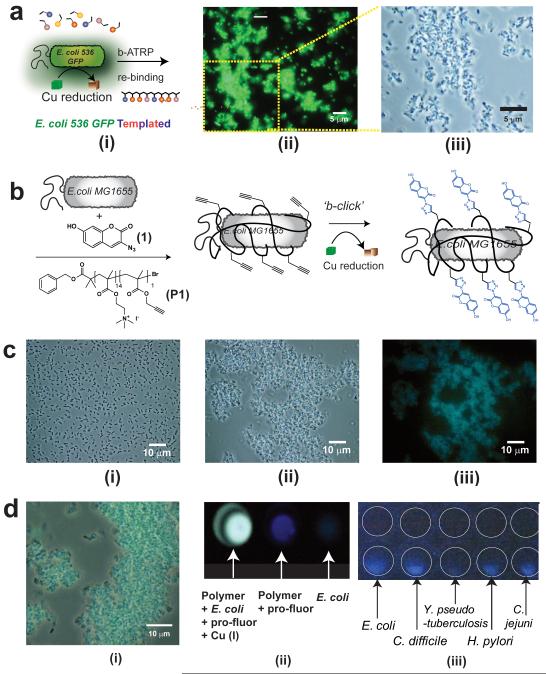
Figure 4. Synthesis in presence of pathogen analogue bacterial strains and in situ labelling of clinical isolates.
Polymers were grown, as shown schematically in (A i), in suspensions of uropathogenic E. coli 536 GFP and the resultant polymer fractions were separated with salt washes as before to recover templated polymer (STP). Micrographs (A) of subsequent binding experiments with E. coli 536 GFP shows pronounced clustering of cells in fluorescence mode (A ii), with size and scale of aggregates apparent in the magnified phase contrast image (A iii). In (B) is shown schematically in situ labelling of cells via bacterial cell-instructed (“b-click”) chemistry. Incubation of non-fluorescent E.coli MG1655 with 3-azido-7-hydroxycoumarin (1) maintains dispersed suspension of cells (C i), while addition of cationic polymer (P1) results in cell clustering (C ii) Reduction of copper at the cell surface leads to no change in cell cluster state, but marked fluorescence in microscopy image (C iii). Efficiency of “b-click” shown in expanded merged phase contrast and fluorescence images (D i) and in the wells of assay plates (D ii). Image capture on a mobile phone camera (D iii) indicates ability to detect pathogenic bacteria including Escherichia coli, Clostridium difficile, Yersinia pseudotuberculosis, Helicobacter pylori and Campylobacter jejuni even in un-optimised conditions. White circles have been added to the image to aid identification of wells, control wells containing bacteria but no labelling components are shown above the wells containing in situ-labelled bacteria.