Figure 2.
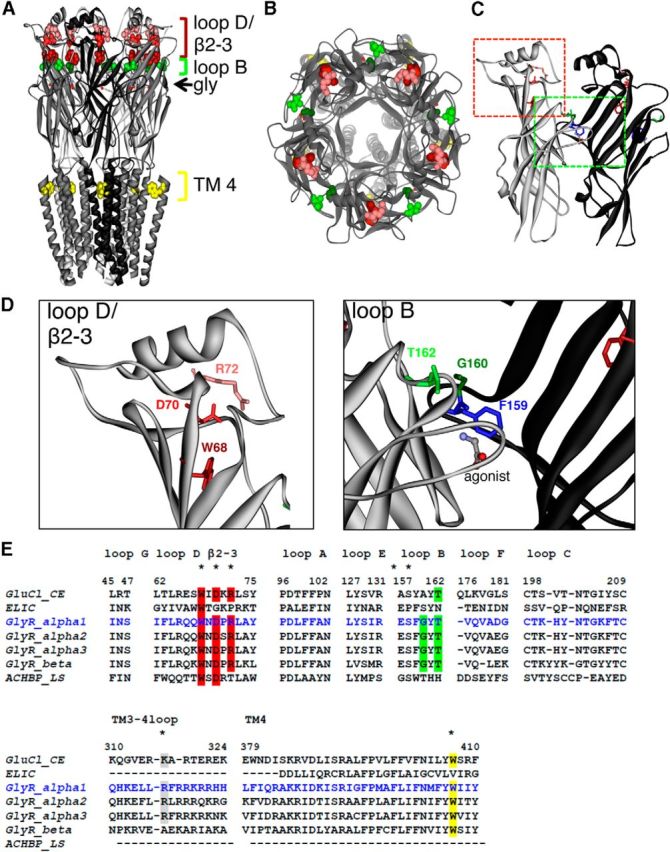
Structural model of GlyR based on GluCl structure. A, View parallel to lipid membrane. The glycine ligands, which are bound at each subunit, are represented in stick form. The amino acids are shown in spacefill representation: W68 (red), D70 (light red), and R72 (pink) are located in loop D/β2–3; G160 (dark green) and T162 (light green) are located in loop B. W407 (yellow) is located in TM4. B, View of the human GlyR looking down the pore axis toward cytosol. C, Dimer interface with colored rectangles pointing to loop B (green) and loop D/β2–3 (red). D, Enlarged view to loop B and loop D/β2–2; affected residues are marked. E, Alignment of various human GlyR sequences of subunits α1 (blue), α2, α3, and β compared with the AChBP from L. stagnalis, ELIC, a prokaryotic homolog of the CLR family, and GluCl from C. elegans. Loop sequences are shown from loops A–G, part of the ICD = TM3–4 loop covering residues 310–324, and TM4. Residues affected in patients with hyperekplexia are highlighted with different colors (red in loop D/β2–3, green in loop B, gray TM3–4 loop, yellow TM4). All mutated residues are highly conserved among sequences shown.
