Figure 4. Micronuclear genome instability in ORC1 knockdown cells.
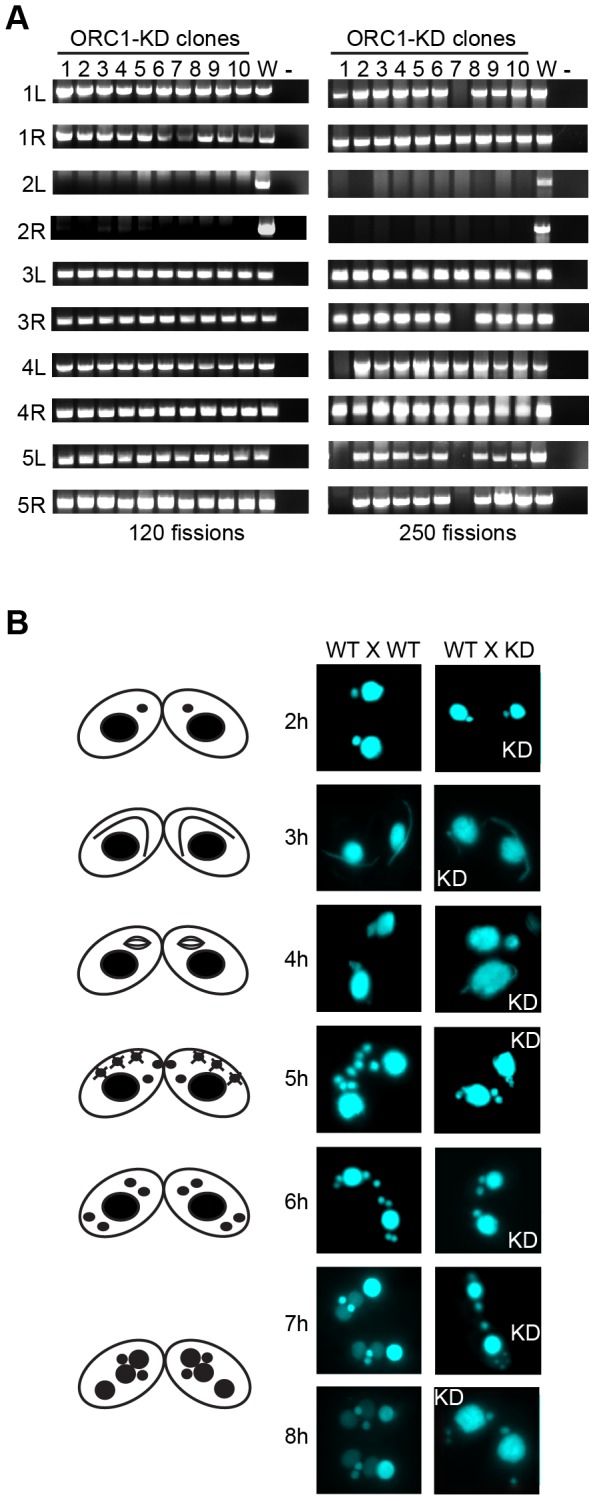
(A) ORC1 knockdown (ORC1-KD) cells were propagated following the establishment of clonal lines. Genomic DNA was isolated at 120 and 250 fissions and subjected to PCR amplification with the primer sets that span sites for chromosome breakage sequence (CBS)-mediated chromosome fragmentation in the developing macronucleus. PCR primers derived from the right (R) and left (L) arms of all five micronuclear chromosomes were tested. 1–10, clonal ORC1-KD; W, wild type strain CU427; (−), PCR reactions in the absence of template DNA. (B) Cytological examination of crosses between wild type strains (CU427 X CU428), or CU427 X ORC1-KD. Nuclei were visualized with the DNA staining dye DAPI. Cartoons depict the progression of wild type mating cells during development. 3 h: micronuclear elongation (crescent); 4 h: micronuclear meiosis; 5 h: four haploid meiotic micronuclei, three of which subsequently undergo programmed nuclear death (PND); 6 h: postzygotic division; 7–8 h: macronuclear anlagen differentiation. Prior to mating, one of the parental strains was incubated with mitotracker red dye to determine the identity of each partner in mating pairs.
