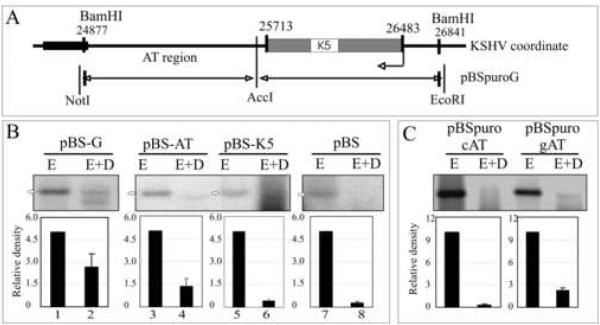
Figure 4.
The AT Region of the G fragment can support replication. (A) Schematic of the G fragment DNA sequence which lies between 24877-25713nt on KSHV coordinates (Russo et al., 1996). (B) Hirt DNA extracted from 293 cells transfected with G, AT or K5 fragments were digested with EcoRI (5%, to linearize) or EcoRI +DpnI and hybridized with 32P labeled ampicilin gene probe. The DpnI resistant bands were quantified. Intensities of input EcoRI lanes were set to 5 and relative intensities of DpnI band in each lane was calculated based on the respective inputs bands after normalizing against the background of individual lane. (C) AT region of the G fragment containing plasmid (pBSpuro gAT) and another similar length (760bp) control AT region of KSHV genome containing plasmids (pBSpuro cAT) were transfected into 293 cells for DpnI sensitivity assay. 10% of the total Hirt DNA was digested with EcoRI and rest 90% with E+D.