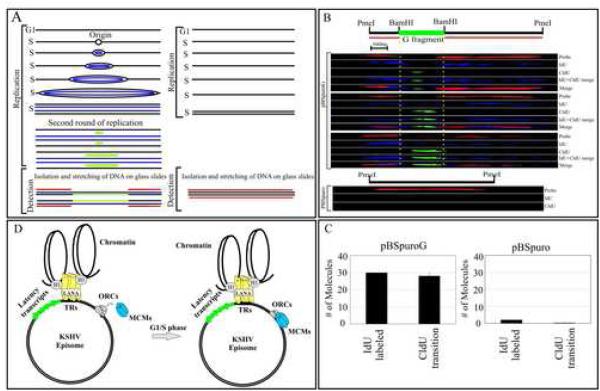
Figure 7.
Single Molecule Analysis of the Replicated DNA. (A) Schematic showing incorporation of nucleotides in DNA containing origin of replication (ori) site. (B) Images of three PmeI linearized pBSpuroG DNA molecules. The hybridization signal detected pBSpuro backbone shown in red. G fragment cloned at BamHI sites of pBSpuro is flanked by regions of pBSpuro shows no hybridization signal. The incorporation of IdU was detected by α-IdU antibody followed by Alexa Flour 350 (blue) showed labeling of the entire length of the molecule after one round of replication. CldU detected by anti-CldU showed the transition site which is most likely the initiation site for the DNA replication. Varying length of CldU labeling was detected in these three molecules. pBSpuro lacking G region did not show incorporation of any halogenated nucleotides due to the lack of replication initiation sites. Hybridization signals detecting pBSpuro vector are shown in red. (C) Graphic representation of the number of molecules showing incorporation of IdU and transition to CldU in pBSpuroG and empty vector, pBSpuro. (D) Proposed model of KSHV persistence with a LANA independent origin of replication. LANA tethers the viral genome to the host chromosome and persists as highly ordered chromatin structure (Stedman et al., 2004; Verma et al., 2006). Binding of ORCs and MCMs at the G region of the KSHV most probably allows binding of other proteins of replication machinery to make pre-RC for latent replication of the KSHV genome.