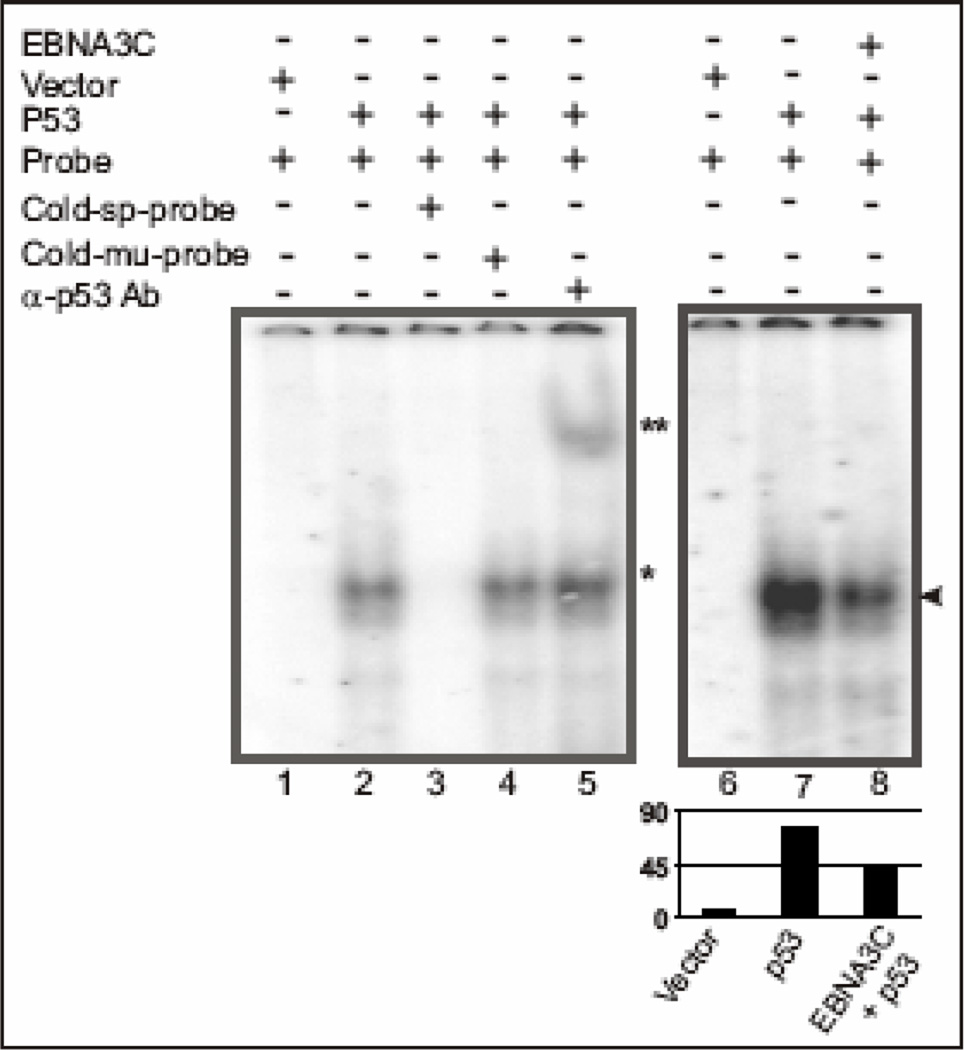
Figure 6.
EBNA3C reduces DNA-binding ability of p53. A probe containing the p53 binding sequence (5’-AGGAAGAAGACTGGGCATGTCTGGGCA-3’) was labeled by Klenow fill-in reaction with [α-32P]dCTP and used for EMSA in presence and absence of EBNA3C. Lanes 1 and 6, probe incubated with nuclear extracts (NE) isolated from SAOS-2 (p53−/−) cells transfected with emprty vector control; lane 2 and 7, probe incubated with myc-tagged p53 expressing NE, showing band shift due to p53 binding (*); lane 3, probe with 200-fold molar excess of cold specific competitor, which abolished completely the p53 specific shift; lane 4, probe with 200fold molar excess of cold mutant probe competitor showing no effect on p53 specific shift; lane 5, probe incubated with p53 expressing NE and monoclonal antibody against p53 (DO-1), which super-shifted the p53 specific probe (**); lane 8, probe incubated with NE expressing both p53 and EBNA3C, reduced the p53 specific band intensity most likely due to formation of a stable inhibitory complex between p53 and EBNA3C in vivo (arrowhead).