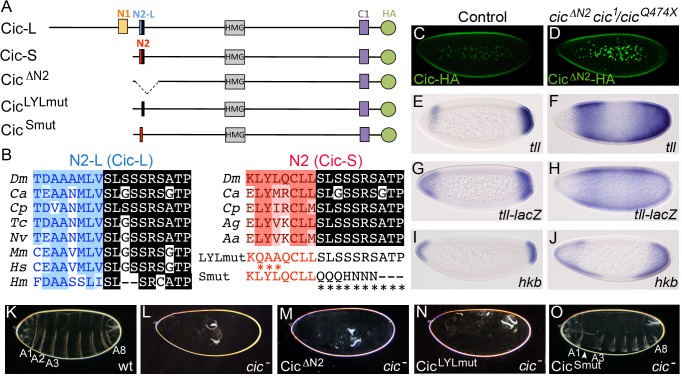
Figure 3. The N2 motif is essential for Cic embryonic function.
(A) Diagram of Drosophila Cic-L and Cic-S isoforms and three derivatives carrying mutations in the N2 motif. Cic-L and Cic-S are generated via use of alternative promoters and splicing sites, which produce different N-terminal domains. At the site of alternative splicing, Cic-L and Cic-S contain two different conserved motifs, N2-L and N2, which include different N-terminal sequences (shown in blue and red, respectively) and a common C-terminal peptide (highlighted in black). Other conserved domains (including the N1 and C1 domains of unknown function) are also indicated. The proteins are shown with an HA tag (green) to allow their visualization in transgenic embryos (see also below). (B) Alignment of Cic N2-L and N2 sequences from different species. Dm, Drosophila melanogaster; Ca, Clogmia albipunctata; Cp, Culex pipiens; Tc, Tribolium castaneum; Nv, Nasonia vitripennis; Mm, Mus musculus; Hs, Homo sapiens; Hm, Hydra magnipapillata; Ag, Anopheles gambiae; Aa, Aedes aegypti. Two different mutations of the N2 peptide (LYLmut and Smut) are also shown below the N2 alignment. (C and D) Expression of Cic and CicΔN2 proteins tagged with the HA epitope in embryos stained with anti-HA antibody; note that both proteins appear downregulated at the embryo poles. (E-M) mRNA expression patterns of tll, tll-lacZ and hkb in wt (E, G, I) and cic mutant (cic1/cicQ474X) embryos expressing CicΔN2 (F, H, J). Cuticle phenotypes of the same genetic backgrounds are shown in K and M, respectively; panel L shows a control cic1/cicQ474X mutant cuticle. (N and O) Cuticle phenotypes of cic1/cicQ474X mutant embryos expressing the CicLYLmut (N) and CicSmut (O) derivatives; only CicSmut rescues the cic phenotype, except for mild segmental defects (arrowhead). A1-A8, abdominal segments 1–8.