Figure 3.
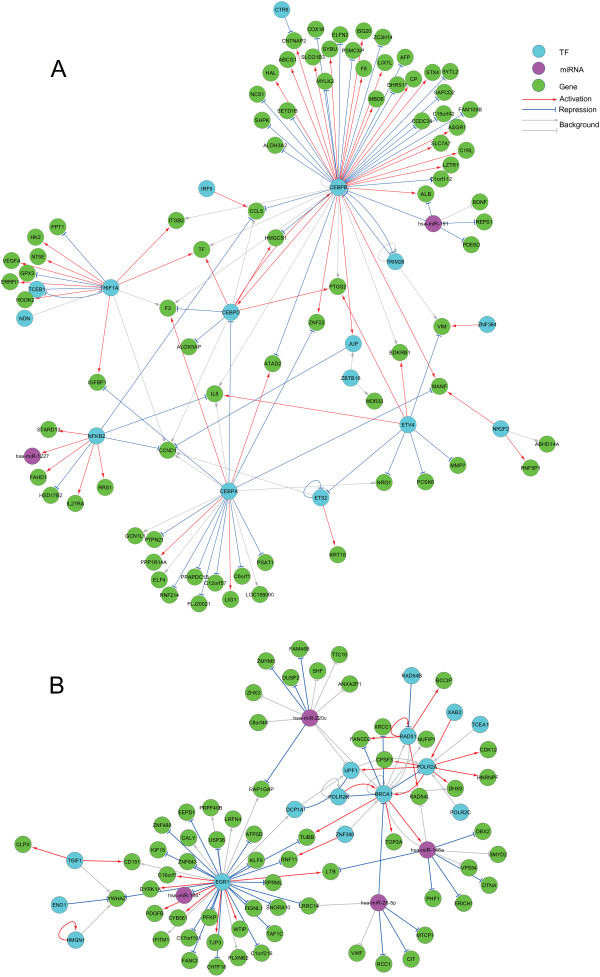
The activated regulatory relationships in two example modules. TFs, miRNAs and genes are in cyan, magenta and green, respectively. The up-regulation and down-regulation are labeled in red and blue, respectively. The background regulatory relationships which are not activated are in gray. The 'arrow’, 'T’ and 'diamond’ shapes of edge terminals denote to up-, down-, and undetermined- regulations, respectively. (A) Module173. This module contains the co-regulation motifs 'CEBPB’-'hsa-miR-191’-'CCL5’ and 'CEBPB’-'hsa-miR-191’-'ALB’/'ISG20’. It also contains some two-node regulatory motifs, e.g., 'ETS2’-'ETS2’, 'NFKB2’-'hsa-miR-1227’, 'IRF5’-'CCL5’ and 'hsa-miR-191’-'REPS1’. (B) Module179. This module contains the regulatory motifs 'BRCA1’-'has-miR-28-5p’-'TUBB’/'POLR2A’, “EGR1’-'hsa-miR-155*’, 'EGR1’-'hsa-miR-146a’-'LTB’ and 'BRCA1’-'hsa-miR-146a’-'PHF1’.
