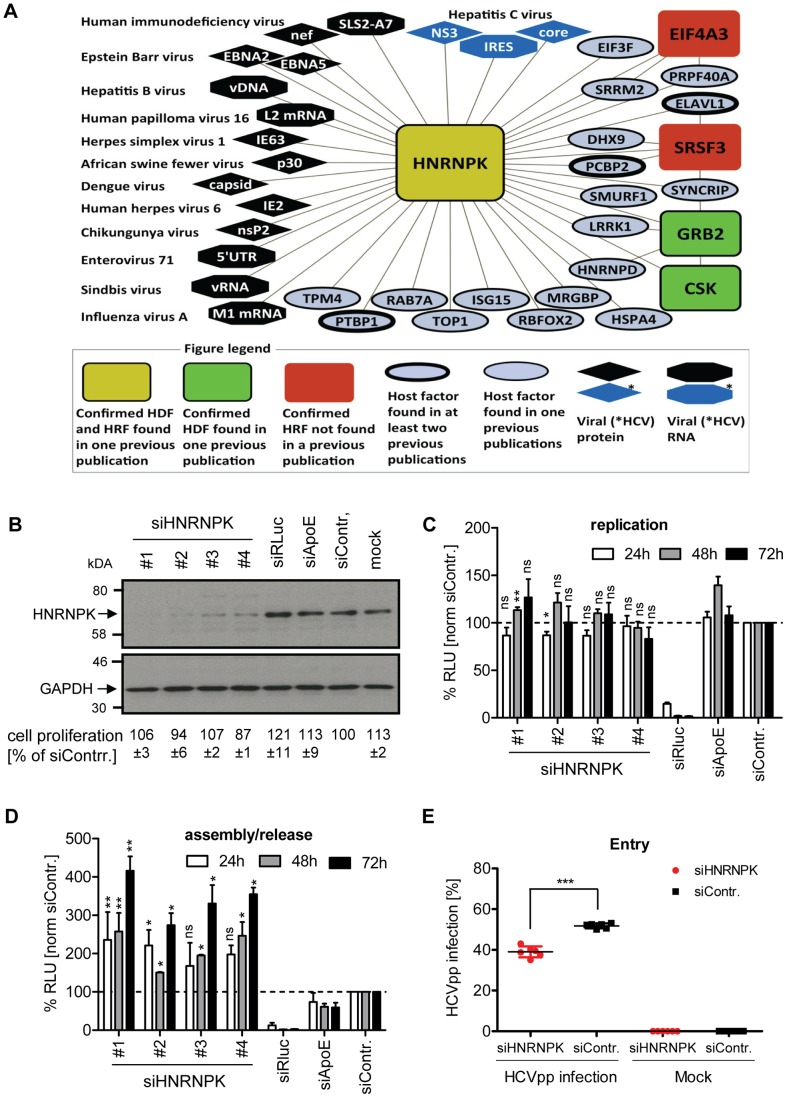
Figure 2. HNRNPK restricts HCV assembly/release but does not affect viral RNA replication.
(A) HNRNPK – virus interaction network. The network shows all human cell proteins (ovals and rectangles), viral proteins (diamonds) and viral RNAs (octagons) and reported interactions with HNRNPK. (B) Silencing efficiency of HNRNPK siRNAs. Huh7.5 cells were electroporated with 2.5 µM of indicated siRNA or mock electroporated 48 h prior to infection with the Renilla luciferase reporter virus JcR2a (MOI = 0.4 TCID50/cell). Cells were harvested 48 h later. Western blot was performed to determine silencing efficiency achieved with siRNAs specified in the top. HNRNPK was detected by using a mono-specific antibody; GAPDH served as loading control. siRNA targeting Renilla luciferase served as positive control for entry/replication assay, siRNA targeting ApoE was used as positive control for assembly/release assay, non-targeting siRNA (siContr.) and mock-treated cells were used as a negative controls. Western Blot represents one of two independent experiments. For HNRNPK silencing, the same siRNA sequences were used as in the validation screen. Quantification of cytotoxicity assays (cell proliferation) using cells transfected with given siRNAs are specified below each lane. Given are mean values ±SD of two independent experiments, normalized to siContr.-silenced cells. (C) Effect of HNRNPK silencing on HCV RNA replication or (D) virus particle production. Huh7-Lunet cells were co-electroporated with 2.5 µM of indicated siRNA and JcR2a in vitro transcripts. Non-targeting siRNA was used as negative control (siContr.). Cells and virus-containing supernatants were harvested at given time points after electroporation. (C) Cell lysates were used for measuring Renilla luciferase activity (relative light units, RLU) to detect effects on HCV replication. (D) Virus-containing supernatants of cells harvested at given time points were used for infection of naïve Huh7.5 cells and Renilla luciferase activity was determined 72 h later to estimate effects of HNRNPK silencing on HCV particle production. All values were normalized to cell viability. Non-targeting control siRNA (siContr.) was set to 100% (dotted line). Bars represent the mean ±SD of three independent experiments. (E) Effect of HNRNPK silencing on HCV entry. Huh7.5 cells were electroporated with 2.5 µM of a mixture of siRNA #1 - #4 targeting HNRNPK or the non-targeting siRNA (siContr.). After 48 h, cells were transduced with YFP-HCVpp (Con1) and the number of infected cells was determined by flow cytometry 72 h later. Non-transduced cells served as negative control. ***, P-value ≤0.0005; **, P-value ≤0.005; *, P-value ≤0.05; ns, non-significant. Statistical analysis was performed by using Student's t-test, referred to the non-targeting control siRNA (siContr.).