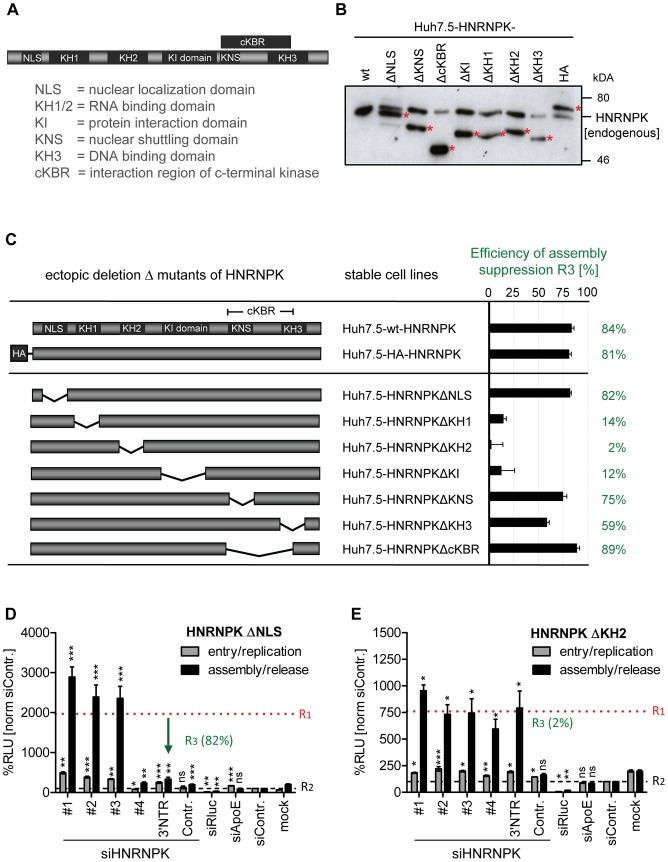
Figure 5. RNA and protein binding domains of HNRNPK are essential for restriction of HCV particle production.
(A) Schematic of HNRNPK protein domains: NLS [aa 21–37]; KH1 [aa 46–98]; KH2 [aa 149–197]; KH3 [aa 391–439]); KI [aa 246–337]; cKBR [aa 338–425]; KNS [aa 338–361]. (B) Stable expression of HNRNPK mutants in Huh7.5 cells. Expression levels of ectopically expressed HNRNPK proteins (labeled with red asterisks), in addition to the endogenous protein (wt), were determined by Western blot. (C) Suppression of HCV particle production by HNRNPK mutants. Huh7.5 cell pools stably expressing HNRNPK proteins specified in the middle column were electroporated with 2.5 µg of siHNRNPK-3′NTR targeting only endogenous HNRNPK. Forty eight hours later, cells were infected with JcR2a (MOI = 0.4 TCID50/ml). R3 values representing the capacity of a given HNRNPK mutant to suppress HCV particle production were calculated as described in the legend to Fig. 4. Examples of results are shown in panel (D) for HNRNPKΔNLS and (E) for HNRNPKΔKH2. Note that the binding region for siHNRNPK#4 is lost because of the NLS deletion. The complete data sets are displayed in S5 Fig. ***, P-value ≤0.0005; **, P-value ≤0.005; *, P-value ≤0.05; ns, non-significant. Statistical analysis was performed by using Student's t-test, referred to the non-targeting control siRNA (siContr.).