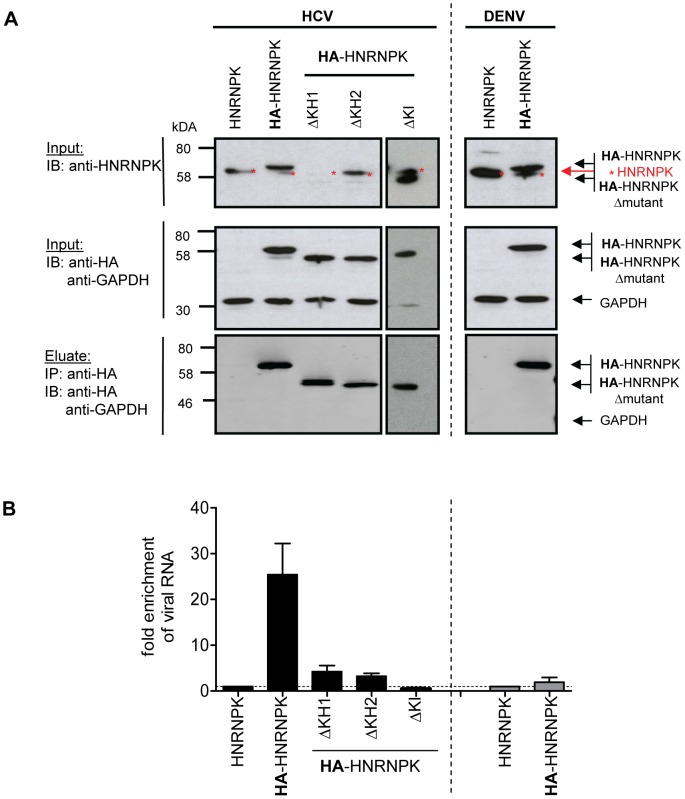
Figure 6. HNRNPK interacts with HCV but not DENV RNA genomes.
(A) Immunoprecipitation of HA-HNRNPK. Huh7.5 cells stably expressing unaltered, HA-tagged HNRNPK, or HA-tagged mutants ΔKH1, ΔKH2 or ΔKI were electroporated with genomic Jc1 (HCV) RNA. Genomic Dengue Virus (DENV) RNA was used as a control. Cells were harvested 48 h post electroporation, lysates were used for HA-specific immunoprecipitation and captured complexes were analyzed by Western blot. Input was analyzed with a HNRNPK and a HA-specific antibody (upper and middle panel, respectively); GAPDH served as loading control. Immunocomplexes (corresponding to 10-fold the amount of the input) were analyzed with the HA-specific antibody (lower panel). Note that the ΔKH1 mutant is not recognized by the HNRNPK-specific antibody, but reacted with the HA-specific antibody. (B) Quantitative analysis of co-precipitated HCV and DENV RNA. Co-precipitated viral RNA was determined by RT-qPCR and normalized to input RNA. Fold enrichment of captured viral RNA was calculated by comparing the HA-tagged to the untagged HNRNPK-fraction (set to 1; dotted horizontal line). Bars represent the mean ±SD of at least three independent experiments.