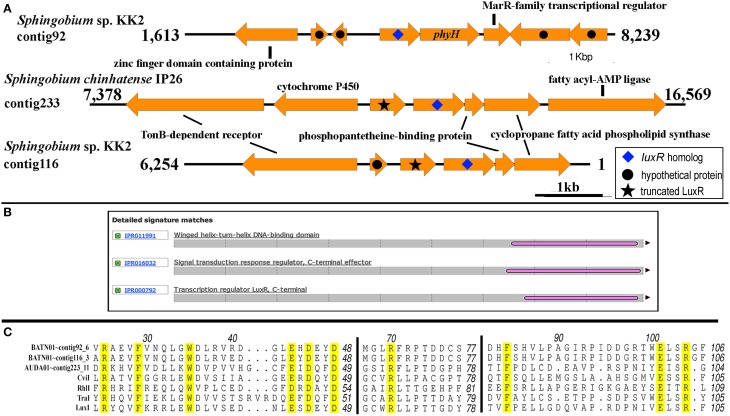
Figure 7.
Genomic and genetic evidence for the presence of luxI solos in sphingomonads. (A) Gene neighborhood showing non-LuxR genes located in the vicinity of the putative LuxI genes (arrow with blue diamond). Analysis of the translated protein sequence for the gene upstream and convergently oriented to the putative luxI gene in Sphingobium chinhatense IP26 and Sphingobium sp. KK2 indicates that it may be an N-terminal truncated LuxR protein (arrow with black star). (B) A representative Interproscan analysis domain analysis of the N-terminal truncated LuxR protein noted in Figure 7A. (C) Protein alignment of the putative LuxI solos. Number above the alignment corresponds to the amino acid residue of TraI. Amino acid residues are conserved in all LuxI-type proteins (Fuqua and Greenberg, 2002) are highlighted in yellow.