Figure 4.
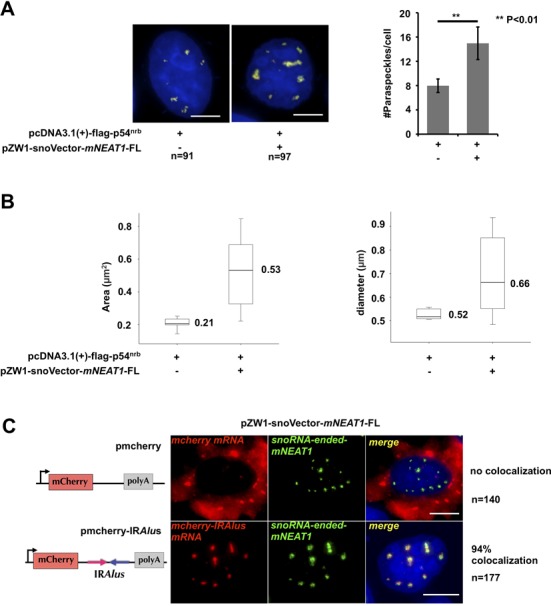
Characterization of mNEAT1 expressed from snoVector. (A) Staining of paraspeckles in cells transfected with or without snoRNA-ended-mNEAT1. Left, HeLa cells were co-transfected with Flag-p54nrb and pZW1-snoVector-mNEAT1-FL or Flag-p54nrb alone, followed by co-staining of Flag-p54nrb (red) and mNEAT1 (green) or endogenous hNEAT1 RNA (green). Both hNEAT1 and mNEAT1 probes were added (green) at the same time to indicate paraspeckles. All nuclei were counterstained with DAPI. Scale bars indicate 5 μm in all panels. Right, quantitative analysis of the average number of paraspeckles per cell by Image pro plus. Ninety-one cells (without sno-ended-mNEAT1) and 97 cells (with sno-ended-mNEAT1) were randomly selected. P values from one-tailed t-test in the pairwise comparison are shown (**P < 0.01). (B) Quantitative analysis of the average area (left) and diameter (right) of individual paraspeckles in cells transfected with or without snoRNA-ended-mNEAT1. Pictures acquired from (A) were analyzed by Image pro plus using the same parameters. (C) SnoRNA-ended-mNEAT1 co-localized with mcherry-IRAlus. HeLa cells were co-transfected with pZW1-snoVector-mNEAT1-FL and pmCherry-C1 or pmCherry-C1-IRAlus containing a pair of inverted repeated Alu elements (IRAlus) in the 3’UTR of mcherry mRNA, followed by co-staining of mcherry mRNA (red) and snoRNA-ended-mNEAT1 (green). One hundred forty cells (transfected with pmCherry-C1) and 177 cells (transfected with pmCherry-C1-IRAlus) were randomly selected.
