Figure 1.
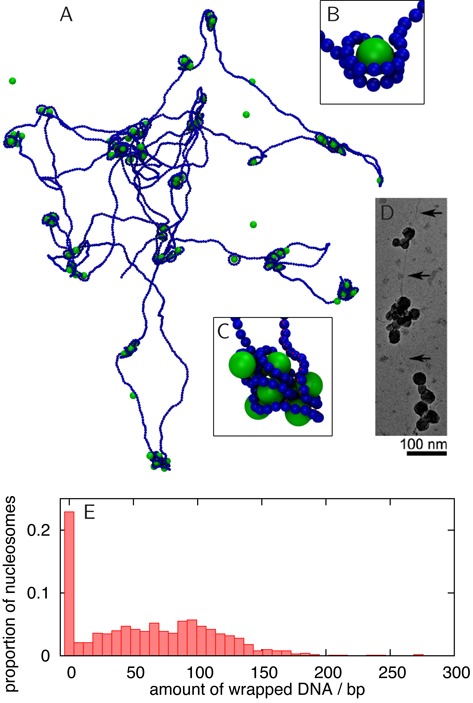
Spherical nanoparticle model synthetic nucleosomes. (A) Snapshot from an MD simulation of a 22.1 kbp DNA molecule interacting with 100 spherical nanoparticles (diameter 6.75 nm), taken 0.18 ms after the system was initialised with nanoparticles positioned randomly and the DNA in an equilibrium configuration. The spheres and DNA interact with an energy 4.3kBT. The in silico reconstitution leads to the formation of nanoparticle–DNA aggregates. (B) Snapshot of a nanoparticle wrapped by two turns of DNA, as in chromatin: this structure is very rare in the simulation. (C) Detailed view of a nanoparticle–DNA cluster. (D) Transmission electron microscopy image of T4 DNA in complex with poly(L-lysine)-coated silica nanoparticles of diameter 15.1 nm (see (25) for details). Clusters of nanoparticles similar to those in simulations are observed. Reproduced from Figure 2f in (25) with permission. ‘Copyright (2005) by the American Physical Society.’ (E) Histogram from simulation data showing how much DNA is associated with each particle, including data from 10 different simulations. A canonical nucleosome will wrap ∼147 bp of DNA in ∼1.75 turns. Clearly, most spheres are associated with an ‘incorrect’ amount of DNA, and indeed the self-assembled structure is very different from a normal chromatin fibre.
