Figure 2.
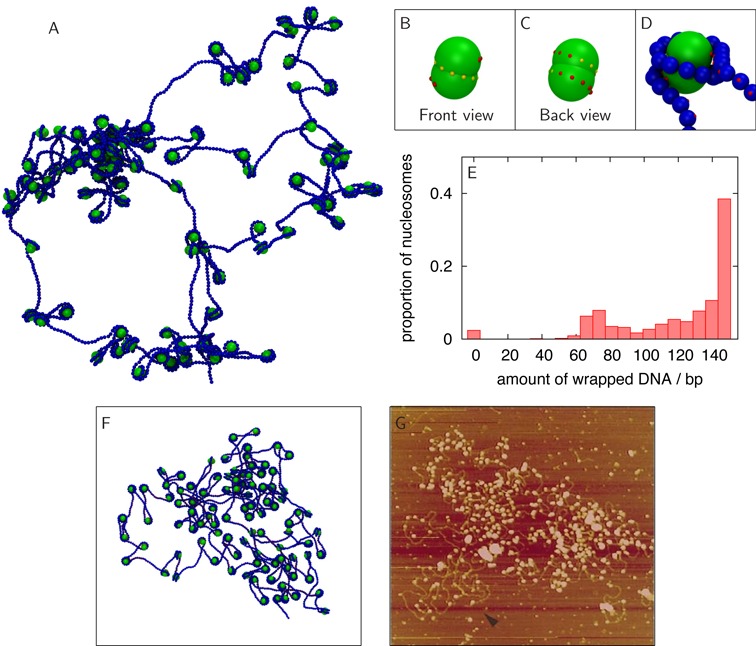
Model nucleosomes with a helical path of binding sites switched on hierarchically form a more realistic chromatin fibre, but with some defects. (A) Snapshot showing a simulation of 100 nucleosome cores and a 22.1 kbp DNA molecule, after 0.3 ms. The image is a projection of a 3D configuration. Initial conditions were to position the cores at regular intervals along a DNA molecule in an equilibrium conformation. (B,C) Each model octamer has a helical binding path which can make contact with the minor groove of the model DNA (Supplementary Figure S1, Supplementary Movie S1). (D) A correctly wrapped nucleosome, with the DNA binding along the helical path on the model histone. (E) Histogram showing how much DNA is associated with each core (i.e. the amount of DNA wrapped), including data from 10 different simulations with identical parameters to that shown in (A). It can be seen that around 40% of the cores are in correctly formed nucleosomes (which wrap ∼147 bp of DNA). (F) Image from a simulation similar to that shown in (A), but here the structure has been flattened onto a plane for better comparison with microscopy images (see Supplementary Data and Supplementary Figure S5 for details). (G) Atomic force microscopy image of a chromatin fibre from a salt dialysis experiment performed on a long DNA loop (see (41) for details). An uneven distribution of nucleosomes is shown, and this could result from defects similar to those found in simulations. Reproduced with permission from Figure 4 in (41).
