Figure 3.
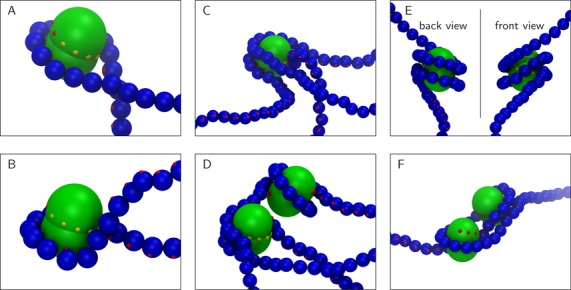
Close-up views of common self-assembly defects found in the simulation presented in Figure 2. (A,B) Partially wrapped nucleosome cores. Full wrapping in (A) would require either enough time for the strands to move around each other (i.e. time for the configuration of the entire DNA molecule to change), or strand passing. In (B) the configuration in (A) has been ‘locked’ due to a further contact between the DNA next to the already wrapped segment and one of the yellow exposed binding patches on the octamer. Resolution of the defect now requires even more time, as the non-canonical binding must first be reversed. (C,D) The exposed binding sites in partially wrapped cores can interact with DNA regions which are far away in sequence; this can lead to loop formation. In (D) this disrupts the wrapping of another nearby nucleosome. (E) DNA wrapping in a right-handed configuration around the core. The DNA does not follow the left-handed path of the core binding sites, and some binding sites remain exposed. (F) Full wrapping can be inhibited if nucleosome cores bind in close proximity along the DNA.
