Figure 5.
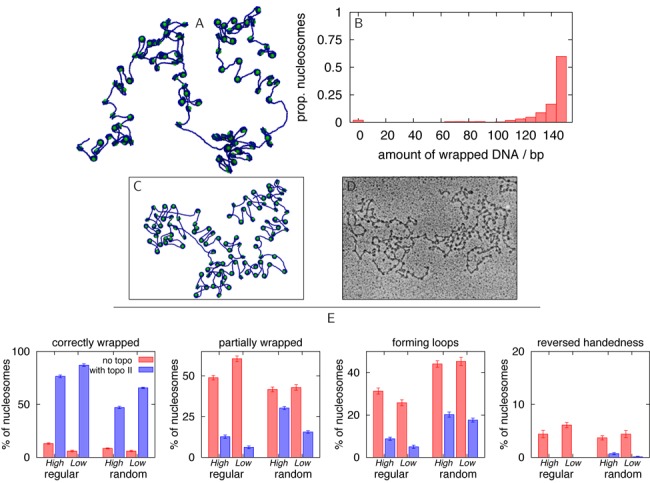
Chromatin self-assembly in the presence of topo-II like action. (A) Snapshot showing a simulation of 100 nucleosome cores and a 22.1 kbp DNA molecule, after 0.3 ms, using the ΔLk = −1.0 model nucleosomes. The image is a projection of a 3D configuration. Initial conditions where the cores are pre-positioned at regularly spaced intervals along a DNA molecule in an equilibrium conformation are used. The action of the enzyme topo-II was modelled by allowing the strands to pass though each other for about 0.1 ms, 0.1 ms after the simulation started (see Supplementary Data). (B) Histogram from simulation data showing how much DNA is associated with each core (i.e. the amount of DNA wrapped), including data from 10 different simulations with identical parameters to that shown in (A). A correctly formed nucleosome will wrap ∼147 bp of DNA in ∼1.75 turns. (C) Image from a simulation similar to that shown in (A), but here the structure has been flattened onto a plane for better comparison with microscopy images. (D) EM image of a native chromatin fibre which has been extracted from chicken erythrocytes, stripped of H1 and H5 linker histones, and treated with tripsin to remove histone tails (see (47) for details). Reproduced with permission from Figure 7c in (47). (E) Histograms from simulation data counting the numbers of correctly formed nucleosomes and defects for the reconstitutions in Figures 2 and 4, and for different initial conditions (regularly or randomly pre-positioned cores), and for high and low nucleosome density (see Supplementary Data for exact values).
