Figure 1.
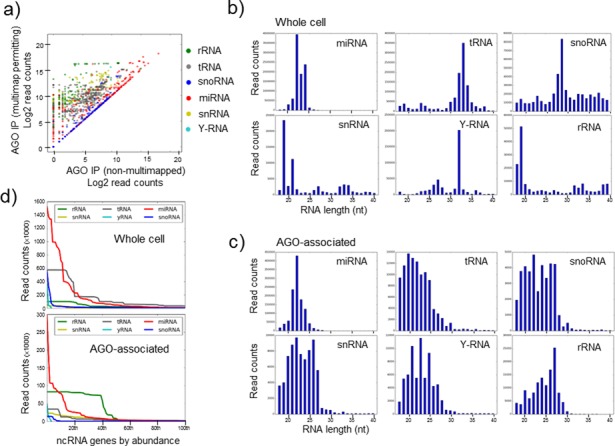
Argonaute binds small RNA from diverse sources. (a) Comparison of the transcript levels of small RNA genes using standard (default) mapping parameters, where multi-mapping reads are proportionately distributed across loci (horizontal axis), to custom parameters that permit multi-mapping (vertical axis) and reads are tallied independently for all individual sequences. For each gene, only the most abundant transcript is shown as a representative (for comparison, all transcripts are represented in Supplementary Figure S1). This highlights genes and small RNA classes whose expression may be under-estimated in standard analyses. (b,c) The relative abundance of transcripts of various lengths derived from diverse sources in both (b) the whole cell and (c) the AGO immunoprecipitate from MDA-MB-231 cells. The vertical axis represents read counts and the horizontal axis the length of the small RNA transcript. (d) For each class of small RNA, the 100 most abundant ncRNA genes for each class are ordered on the horizontal axis according to their abundance, which is plotted as read count on the vertical axis. This reveals both the predominant association of a subset of miRNAs with AGO and the association of small RNAs derived from other sources which individually may be in excess of many lowly and moderately expressed miRNAs.
