Figure 5.
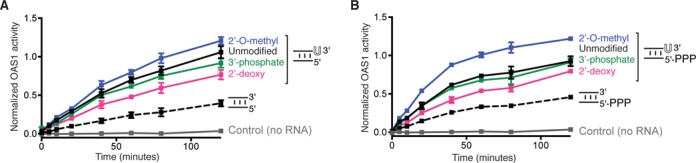
3′-end modifications have minor effects on 3′-ssPy activity and are relatively unaltered by 5′-ppp on the reverse strand of 18-bp RNA. (A) Assays of OAS1 activation by dsRNA duplexes with chemical modifications to the 3′-ssPy motif ribose group: 2′-O-methyl, 2′-deoxyribose and 3′-phosphate. dsRNA with unmodified 3′-ssPy (one single-stranded 3′-end uridine) and without a 3′-ssPy motif are shown for comparison. None of the modifications reduced activity to the level of the dsRNA lacking a 3′-ssPy motif but subtle changes in activation suggest an influence of the ribose sugar pucker on 3′-ssPy motif activity. (B) As panel (A) but for dsRNA duplexes with a 5′-triphosphate group on the complementary strand. The 5′-triphosphate group did not alter relative effects of the modifications on 3′-ssPy motif potency but does appear to modestly enhance activation by the 3′-ssPy motif with 2′-O-methyl ribose modification. In all panels data are normalized to the 18-bp dsRNA with the unmodified 3′-ssPy motif.
