Figure 2.
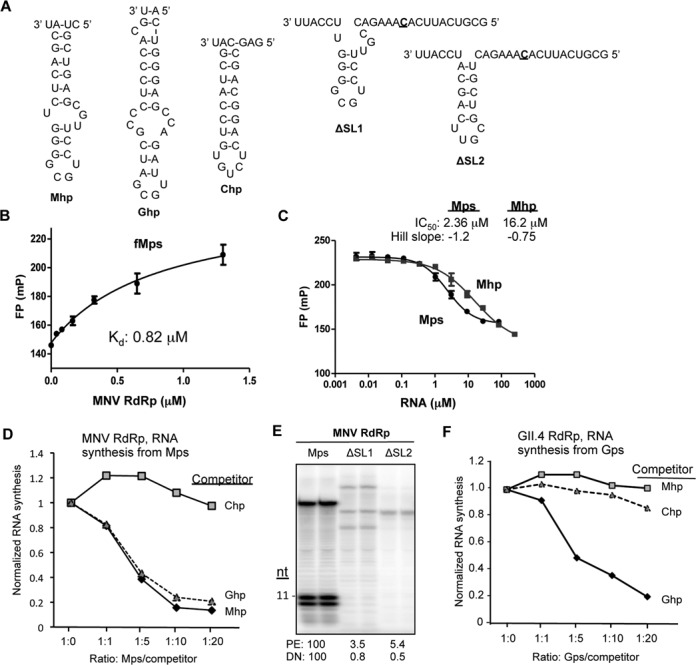
The hairpin within the MNV subgenomic proscript is required for binding by RdRp and to direct RNA synthesis. (A) RNAs use in this analysis. The names of the RNAs are in bold. ΔSL1 and ΔSL2 have deletions in the hairpin of Mps, but the template sequences are unchanged. The template initiation cytidylate is in bold and underlined. (B) Fluorescence polarization results assessing the affinity of MNV RdRp for fMps. RNA fMps was chemically synthesized to contain a 5′ 6-carboxyfluorescein (6-FAM) attached to Mps. The binding isotherm is shown and all data points were assessed in triplicate. The results are reproducible with four independent assays. (C) A competition assay assessing the inhibitory activities of Mps and Mhp on MNV RdRp binding to fMps. The effective inhibitory concentration (IC50) and the Hill slope for the competitor RNAs, Mps and Mhp, are shown above the graph. IC50 values were derived using the algorithm developed by Nikolovska-Coleska et al. (61). (D) The hairpin in Mps and the Gps can bind to the MNV RdRp. The graph shows the results from an RNA synthesis assay performed with the MNV RdRp in the presence of increasing concentrations of three competitor RNAs, Chp, Ghp and Mhp. Comparable results were obtained in three independent experiments. (E) Deletions in the hairpin within Mps severely reduced de novo initiated RNA synthesis. The gel image was from an RNA synthesis assay with the template RNAs all being at 0.05 μM. (F) The GII.4 norovirus RdRp can specifically recognize the hairpin in the GII.4 norovirus proscript. The graph shows the results from RNA synthesis assay performed with the MNV RdRp in the presence of increasing concentrations of three competitor RNAs, Chp, Ghp and Mhp.
