Figure 5.
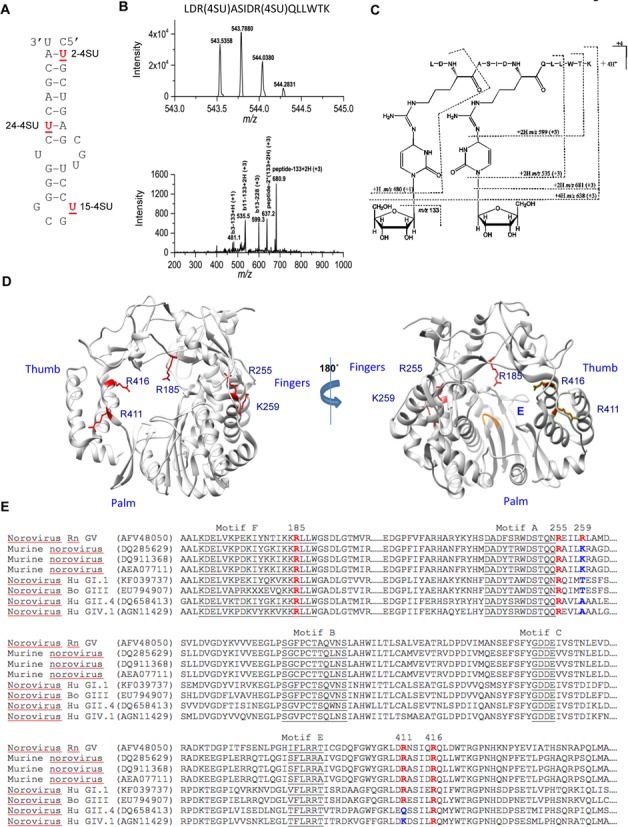
Residues of the MNV RdRp that contact 4-thiouridylate (4SU) within the Mhp. (A) Locations of the 4-SU within the chemically synthesized Mhp RNAs. The names of the RNAs with each of the 4-SU residue are in bold. All samples were processed by a UV crosslinking protocol that included exhaustive digestion of the RNAs that should leave only the 4-SU nucleotide covalently attached to peptides. (B) Mass spectrum (MS) and collision-induced dissociation (CID) spectrum of the peptide ion [LDR(4SU)ASIDR(4SU)QLLWTK- S2]4+ obtained on a Thermo LTQ-Orbitrap instrument. (Top) High-resolution mass spectrum (MS) of the peptide ion LDR(4SU)ASIDR(4SU)QLLWTK- S2]4+ (RdRp residues 409–422) from the reaction with 15-2SU substituted RNA. The measured monoisotopic mass (5 ppm mass error) is at m/z 543.5358 (4+). (Bottom) CID spectrum of this peptide ion at m/z 543.5. The CID fragment ion assignments are illustrated with the dashed lines in (C) structure. (C) Proposed structure and collision induced dissociation (CID) fragments (dashed lines) of the doubly-substituted 4SU peptide. The most common loss upon fragmentation is that of the ribose sugar (m/z 133) from either the parent ion or its ‘b ion’ peptide fragments. (D) A summary of the locations of the residues in the MNV RdRp that were identified to contact the three 4-SU-modified RNAs. The side chains of the residues are shown in red. (E) Alignments of the sequences of the norovirus RdRps that are relevant to the residues found in the MNV RdRp which contacted different regions of Mhp. All sequences are shown with their Genbank accession number. The residues in specific motifs conserved in viral RdRps are underlined. The residues identified to contact the 4SU-modified MNV are in red, as are the comparable residues in other norovirus RdRps. Where there are amino acid substitutions in the key residues, the changes are shown in blue.
