Figure 7.
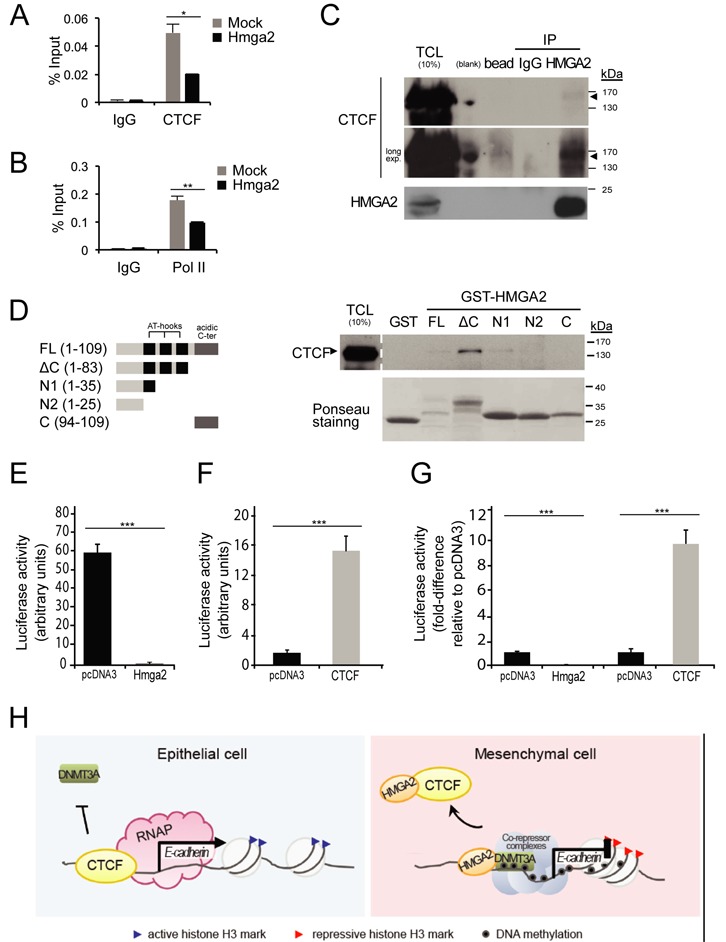
Impact of HMGA2 on CTCF binding to chromatin. (A, B) ChIP-qPCR analyses of CTCF and RNA polymerase (Pol II) on the Cdh1 promoter in NM-Mock and NM-Hmga2 cells. (C) Immunoprecipitation of endogenous HMGA2, followed by immunoblotting for endogenous CTCF in HEK293T cells. IP, immunoprecipitation; TCL, total cell lysate; long exp., long exposure. Arrowheads mark specific protein bands. (D) Purified GST or GST-HMGA2 and its deletion mutants, as schematically depicted in the left panel, were used in a pull-down assay with HEK293T cell extracts and immunoblotted for endogenous CTCF. Input of GST fusion proteins used in the pull-down were visualized by Ponceau-S staining. FL, full-length; ΔC, deletion of C-terminal; N1, N-terminal 1; N2, N-terminal 2; C, C-terminal; TCL, total cell lysate. (E, F) Luciferase reporter assays of human Cdh1 promoter in NMuMG cells co-transfected with empty vector pcDNA3 or pDNA3-HA-HMGA2 (E) or pcDNA3-Flag-CTCF (F). Each bar represents mean ± SD values from triplicate samples. ***P < 0.001, compared to empty vector-transfected cells. (G) The data of panels E and F are re-plotted on a single graph after normalization of the empty vector pcDNA3 conditions to 1, for better comparative visualization. (H) CTCF prevents the spread of heterochromatin and DNA methylation, and promotes E-cadherin transcription by RNA polymerase II complexes (RNAP) in epithelial cells. HMGA2 interacts with CTCF such that CTCF can no longer bind efficiently and protect the Cdh1 region from repressive complexes, e.g. DNMT3A, which enforces methylation of DNA at the locus. Additional co-repressor complexes may be recruited to the locus. The key EMT-TFs, such as Snail, that initiate Cdh1 repression, and which cooperate with HMGA2 are not shown for simplicity.
