Figure 1.
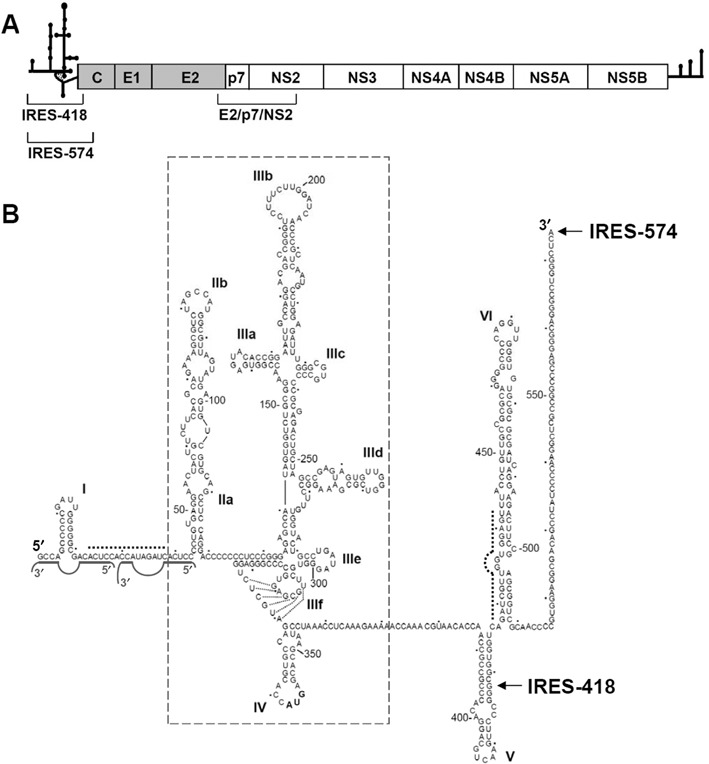
HCV IRES in its natural sequence context. (A) Schematic representation of the 9.6-Kb-long, ssRNA HCV genome. The secondary structures of the 5′ UTR and 3′ UTR are schematized. The structural (gray background) and non-structural (white background) proteins within the viral polyprotein are boxed. The regions corresponding to the IRES-418, IRES-574 and E2/p7/NS2 transcripts are shown. (B) Sequence and secondary structure of the 5′ UTR of HCV, previously deduced by different bioinformatic, biochemical and biophysical methods. Nucleotides are numbered every 50 residues, and domains/subdomains are labeled from I to VI. HCV IRES domains II to IV are boxed. The last residues of the IRES-418 and IRES-574 molecules used in this work are marked with arrows. Dashed lines show the complementary sequences at the I-II spacer (nts 24–38) and the basal region of domain VI (nts 428–442), while the two miR-122-binding sites are marked with solid gray lines.
