Fig. 5.
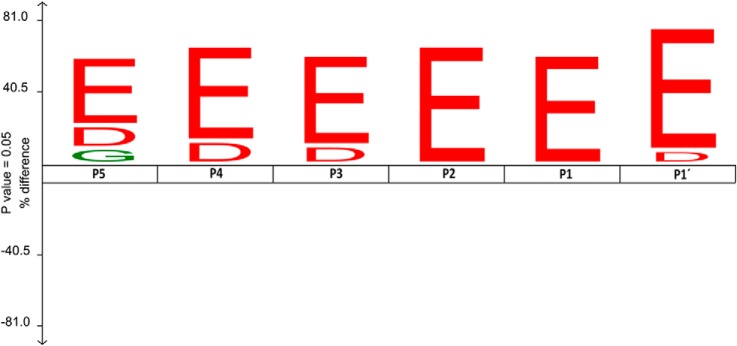
Substrate specificity profile derived from putative CCP1 substrates. IceLogo (35) was used to show the enriched residues present at the different identified CCP1 putative substrate positions as compared with the reference set (i.e. C termini of human proteins in the Swiss-Prot database). For the iceLogo preparation we took into consideration that MCPs sequentially release amino acids. As a result, when a few amino acids are released to get to the final identified product, intermediate products of digestion act as new substrate and are further digested by CCP1. Consequently, intermediate products of digestion were considered as different individual substrates when preparing the iceLogo and, as a result, 35 CCP1 substrates were considered. Only those substrates identified by proteomics were considered here. In the representation the substrate residues are depicted according to Schechter & Berger nomenclature. The frequency of the amino acid occurrence at each position in the sequence set was compared with the occurrence in the reference set. Only statistically significant residues with a p value ≤ 0.05 are plotted. Amino acids height shows the degree of difference in the positional amino acid frequencies in the experimental set as compared with the reference set. Residues that are statistically over- or underrepresented in the experimental set are shown in the upper or lower part of the iceLogo respectively.
