Table I. CSPGs identified in the present study with an intracellular distribution.
| Protein namea | Peptide sequenceb | Glycan modificationc | Sample type |
|---|---|---|---|
| Intracellular granules CSPGs | |||
| Bone marrow proteoglycan | 53ELEEEEEWGSGSEDASK69 | 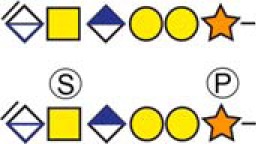 |
Urine |
| aCholecystokinin | 21QPVPPADPAGSGLQR35 |  |
CSF and Urine |
| Chromogranin-A | 419KEEEGSANR427 | 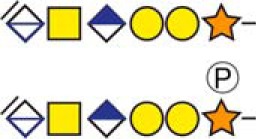 |
CSF and Urine |
| aCollagen and calcium-binding EGF domain-containing protein 1 | 382DLGSGDDHPR391 | 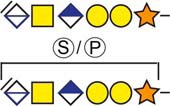 |
Urine |
| aDermcidin | 20YDPEAASAPGSGNPCHEASAAQK42 | 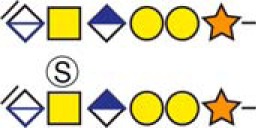 |
Urine |
| aNeuropeptide W | 120APEPALEPESLDFSGAGQR138 | 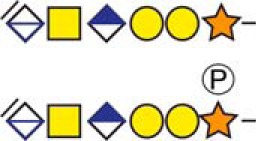 |
CSF and Urine |
| aSecretogranin-1 | 88DPADASEAHESSSR101 | 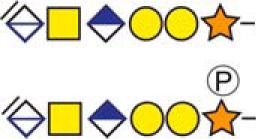 |
CSF and Urine |
| aSecretogranin-1 | 235SSQESGEETGSQENHPQESK254 |  |
CSF |
| aSecretogranin-3 | 35ELSAERPLNEQIAEAEEDK53 |  |
Urine |
a Indicate novel CSPGs identified in this study.
b Bold and underlined serine residues depict established attachment sites while bold depict probable attachment sites.
c The CS-hexasaccharide were identified either without or with sulfate (SO3−) and/or phosphate (PO3−) modifications and their positions on the hexasaccharide structure are indicated. The positioning and distinction of sulfate- (79.9663 Da) and phosphate (79.9568 Da) modifications were made by manually evaluating the MS2-spectra. When the expected modifications could not be identified a bracket including the whole hexasaccharide structure is presented as the expected modification could be, in theory, anywhere on the glycan.
