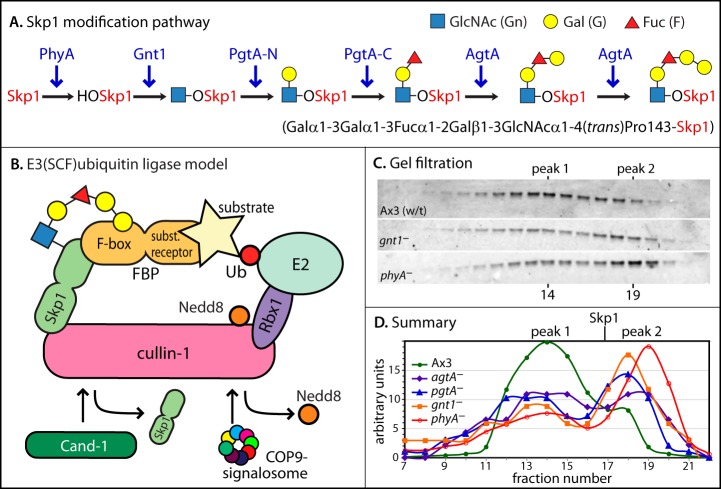
Fig. 2.
Skp1 modification pathway and global analysis of Skp1 interactions. A, Skp1 is sequentially modified by the indicated enzymes (in blue), resulting in the formation of a pentasaccharide at Pro143. B, model of the SCF complex in the context of the overall E3 Ub ligase, from studies in yeast, plants, and animals. Catalysis involves transfer of Ub from an exchangeable Ub-E2 conjugate to the substrate. Removal of Nedd8 by the COP9 signalosome facilitates binding of Cand1 to Cul1, which inhibits binding of Skp1 to Cul1. C, D, vegetative (growth stage) cells were filter-lysed, and a cytosolic fraction prepared via ultracentrifugation was chromatographed on a Superose 12 gel filtration column. Fractions were analyzed via Western blotting (representative examples are shown in C) followed by densitometry (D). The elution position of free Skp1 from a separate trial is indicated.