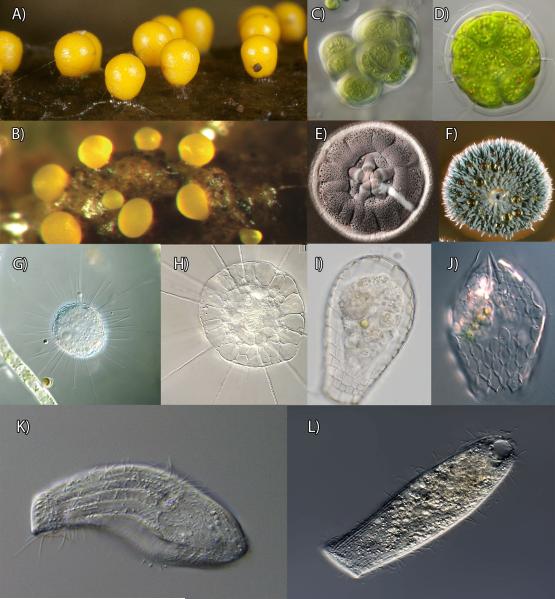
Figure 3.
Examples of striking morphological convergence between bacterial and eukaryotic domains (A-F) and among deeply-diverged eukaryotic clades (G-L). A: Fruiting bodies of Trichia varia, an amoebozoan eukaryote. B: Fruiting bodies of Myxococcus xanthus, a bacterium. C: Gloeocapsa sp., a cyanobacterium. D: Pandorina morum, a green alga. E: a colony of the bacterium Streptomyces. F: a colony of the fungus Penicillium. The last common ancestor between the organisms in figure A and B, C and D, and E and F is the last common ancestor of all life, thus, each of these pairs of organisms must have diverged over 3.5 billion years ago; G: Acanthocystis penardi, the “sun-animalcule”, a centrohelid eukaryote (group with unknown affinities). H: Actinosphaerium eichorni, another “sun-animalcules”, however, molecular data has shown that this is a stramenopile (related to diatoms). I: Quadrullela, an arcellinid amoebozoan, or lobose testate amoeba. J: Euglypha, and euglyphid rhizarian, or filose testate amoeba. K: Stephanopogon apogon, a heterolobosean, the heterolobosea are typically characterized by amoeboid forms with a flagellate life-cycle stage. L: Spathidium, a ciliate. In these eukaryotic pairs of organisms, each of the pair belongs in a very distant clade to the other, with a last common ancestor only billions of years ago. Image credits: B, E, F are from Wikimedia; A, C, D, J are from micro*scope; G, H were kindly provided by Mr. Wolfgang Bettighoefer; J is from Daniel Lahr.