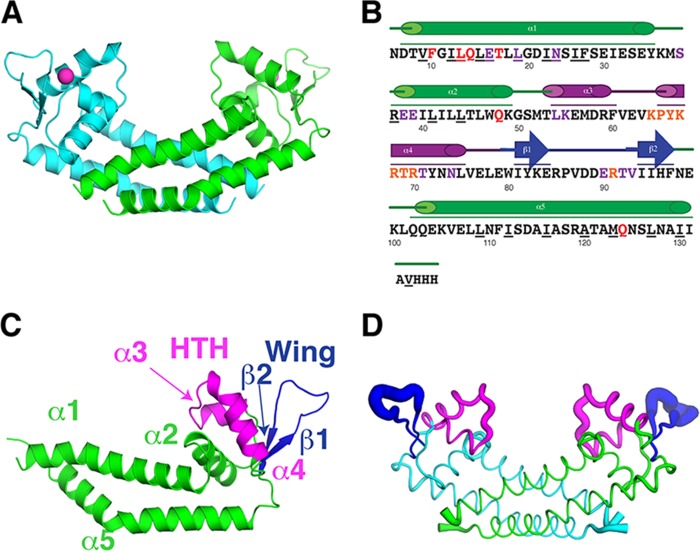
FIG 1.
Rot structure. (A) The Rot dimer is shown in a ribbon representation from a side view. Monomers are colored in either cyan or green. The chloride ion is represented as a magenta sphere. (B) The sequence of the region in the crystal structure from residues 6 to 133 plus three residues from the 6×His tag is shown with the secondary structure elements, including five α-helices and a two-stranded β-sheet. The structural motifs are color coded: the dimerization core helices (α1, α2, and α5) in green, the helix-turn-helix (HTH) (α3 and α4) containing the recognition helix (RH) (α4) in magenta, and the wing (β1 and β2) in dark blue. The underlined residues make dimerization contacts. Red residues are predicted to make contact with protein partners, purple residues are predicted to make nonspecific contact with DNA, and orange residues are predicted to make specific contact with DNA. (C) Structural motifs listed in panel B are shown on the Rot monomer. (D) The Rot dimer is shown in a B-factor putty representation where the thickness of a region is proportional to its local B-factor and thus its flexibility.