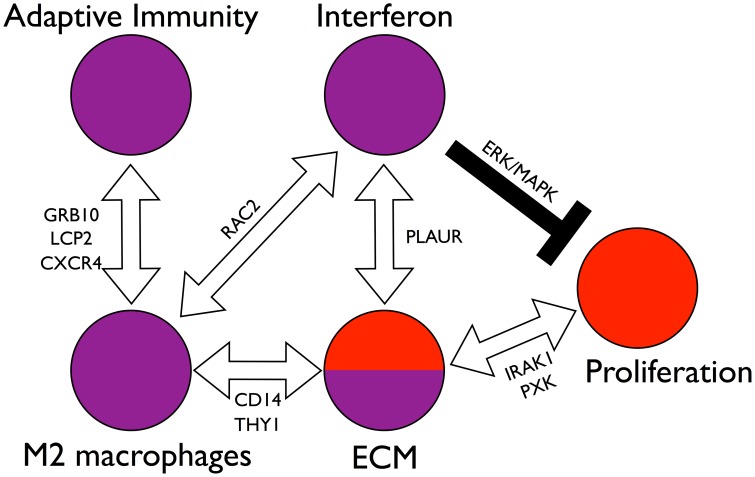
Figure 7. Model of interactions among the components of the network.
The molecular network of Fig. 4 is densely interconnected, implicating many possible interactions between the core molecular processes (interferon activation, M2 macrophage activation, adaptive immunity, ECM remodeling, and cell proliferation). Stepping back from the granular detail of single genes, we see a system of distinct parts through which SSc could be initiated and maintained. Among these are paths of particular interest. The interferon subnetwork and the M2 macrophage subnetwork are connected by RAC2. The M2 macrophage subnetwork in turn is connected to the ECM subnetwork through paths through CD14 and THY1. Suggesting macrophages may influence or drive ECM abnormalities in skin. The interferon subnetwork and the ECM subnetwork are connected through paths containing the pleiotropic and polymorphic gene PLAUR. The M2 macrophage subnetwork is connected to the adaptive immunity subnetwork through several distinct sets of paths through the genes GRB10, LCP2, and CXCR4. The ECM subnetwork is connected to the cell proliferation cluster through TGFβ pathway genes and paths containing the polymorphic genes IRAK1 and PXK, which suggests that ECM remodeling modulates cell proliferation through the TGFβ pathway. The interferon node may negatively regulate proliferation via the ERK/MAPK pathway resulting in the general mutual exclusivity of the inflammatory and fibroproliferative subsets. Thus we see a set of interconnected, balancing feedback loops that can enforce subset homeostasis, but also allow for patients to transition between the subsets, possibly in response to therapy.