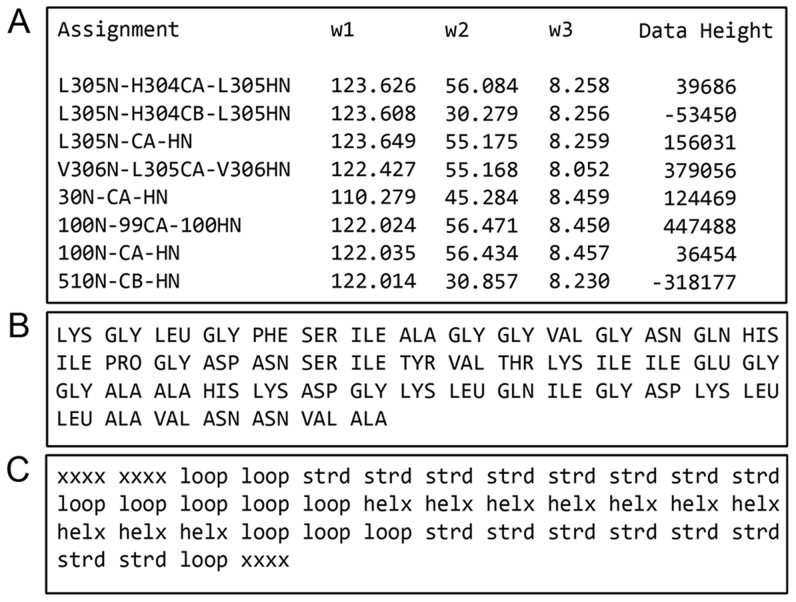
Figure 3. Examples of input files of the supported formats for COMPASS usage.
(A) The HNCACB peak list file exported from Sparky with the column ‘Data Height’, indicating peak intensity. (B) The protein sequence file contains amino acids in uppercase three letter code separated with whitespace. (C) The secondary structure file must contain exactly one secondary structure element for each amino acid in the protein sequence file. The format is ‘xxxx’ for unknown secondary structure element, ‘helx’ for α-helix structure, ‘strd’ for β-strand structure and ‘loop’ for random coil structure written in lowercase letters and separated with whitespace.