Abstract
Polybrominated diphenyl ethers (PBDEs) are flame retardants that have been used in consumer products and furniture for three decades. Currently, very little is known about their fate in the environment and specifically about their susceptibility to aerobic biotransformation. Here, we investigated the ability of the polychlorinated biphenyl (PCB) degrading bacteria Rhodococcus jostii RHA1 and Burkholderia xenovorans LB400 to transform mono- through hexa-BDEs at ppb levels. We also tested the PBDE transforming abilities of related strain Rhodococcus sp. RR1 and the ether-degrading Pseudonocardia dioxanivorans CB1190. The two PCB-degrading strains transformed all of the mono- through penta-BDEs and strain LB400 transformed one of the hexa-BDEs. The extent of transformation was inversely proportional to the degree of bromination. Strains RR1 and CB1190 were only able to transform the less brominated mono- and di- BDE congeners. RHA1 released stoichiometric quantities of bromide while transforming mono- and tetra-BDE congeners. LB400 instead converted most of a mono-BDE to a hydroxylated mono-BDE. This is the first report of aerobic transformation of tetra-, penta- and hexa-BDEs as well as the first report of stoichiometric release of bromide during PBDE transformation.
Introduction
Polybrominated diphenyl ethers (PBDEs) are a class of flame retardants that have been used for more than three decades in a wide variety of manufactured materials such as foams, textiles and plastics at up to 20% by weight (1). PBDEs were widely integrated into products as mixtures of penta-BDEs (mostly penta- and tetra-BDE congeners), octa- BDEs (mostly octa- and hepta-BDE congeners) and deca-BDE. As of 2006, the penta- and octa-BDE technical mixtures have been banned and removed from production in Europe and the Unites States. Some PDBEs are toxic and others cause endocrine disruption in rats and mice (2). As a consequence of their extensive long-term use in consumer products, PBDEs have been detected in a wide variety of environmental samples, ranging from air, water, soil and sediment to biota (1,3–5). Concentrations of total PBDEs in the San Francisco Bay in 2002 ranged up to 0.5 ng/L in water and 212 μg/kg dry weight in aquatic sediments (5). In 2000, PBDE concentrations in human breast milk were reported to be doubling every five years, creating substantial concern about the fate and transport of these compounds and human exposure to them (6).
PBDEs have been shown to be anaerobically and aerobically transformed by microorganisms. Recent work on anaerobic biodegradation has shown that PBDEs are debrominated to less brominated congeners by a variety of anaerobic dehalogenating bacteria (7–9). However, currently very little is known about aerobic biotransformation of PBDEs. In the early 1990s, Schmidt and colleagues isolated two Sphingomonas sp. strains capable of breaking down mono- and di- BDE congeners. Sphingomonas sp. SS3 was capable of transforming and growing on 4-bromodiphenyl ether while strain SS33 transformed 4,4′-dibromodiphenyl ether but was not able to use it as a growth substrate (10,11). Several transformation products were identified, including 4-bromophenol, 4- bromocatechol and bromide. Subsequently, the white-rot fungi Trametes versicolor was shown to convert 4-bromodiphenyl ether to its hydroxylated analog (12). More recently, additional Sphingomonas species have been shown to aerobically break down a number of mono-, di- and tri-BDEs. Sphingomonas sp. PH-07, which grows on diphenyl ether, was shown to transform but not grow on 4-bromo-, 2,4-dibromo-, 4,4′-dibromo and 2,4,4′-tribromodiphenyl ethers (13). In that study, brominated phenols were identified as PBDE breakdown products and in the case of 4,4′-dibromodiphenyl ether, the metabolite 2′3′-dihydroxy-4,4′-dibromodiphenyl ether was also identified.
More highly brominated congeners such as tetra- and penta-BDEs—the most frequently detected congeners in the environment—have not yet been shown to be aerobically biotransformed. Understanding whether bacteria can aerobically transform these congeners is important for understanding the fate of these compounds in the environment. We hypothesized that the chemical similarity of polychlorinated biphenyls (PCBs) and PBDEs would result in similar bacterial transformation patterns. PCB congeners with three or fewer chlorines are susceptible to transformation by certain species of aerobic bacteria resulting in the accumulation of metabolites such as chlorobenzoates. Degradation of PCBs with four to six chlorines has also been observed (14–17). In this study, we followed the disappearance of PBDEs and formation of bromide and other products when two known PCB-degrading bacteria, Rhodococcus jostii RHA1 and Burkholderia xenovorans LB400 and two related strains were exposed to PBDE congeners ranging from mono- to hexa-BDEs. Although experiments in which pure cultures of bacteria are exposed to relatively high concentrations of PBDEs may not be representative of environmental conditions, they provide insight into whether aerobic bacteria are capable of transforming tetra-, penta- and hexa-BDEs.
Materials and methods
Bacterial Cultures
The strains used in this work are Rhodococcus jostii RHA1, provided by Dr. Bill Mohn at the University of British Columbia, Vancouver, Canada (18); Rhodococcus sp. RR1, which was previous isolated from a toluene-contaminated site (19); Burkholderia xenovorans LB400 (DSMZ #17367); and Pseudonocardia dioxanivorans CB1190, supplied by Dr. Rebecca Parales at the University of California, Davis. The Rhodococcus and Pseudonocardia cultures were grown in a mineral medium modified from (20)’ containing: 10 g/L NaNO3, 1.7 g/L K2SO4, 0.37 g/L MgSO4*7H2O, 0.086 g/L CaSO4*2H2O, 5.3 g/l KH2PO4, 10.6 g/L K2HPO4, 5.74 mg/L ZnSO4*7H2O, 3.38 mg/L MnSO4*H2O, 1.24 mg/L H3BO3 and 5 mg/L CuSO4*5H2O. The following ingredients were added after autoclaving: 0.94 mg/L CoMoO4*H2O, 0.22 mg/L FeSO4*7H2O, 0.1 mL/L H2SO4. LB400 was grown in Brunner mineral medium (DSMZ medium # 457). Because the Brunner medium contains chloride and consequently some bromide impurities, LB400 was grown in the same minimal medium as RHA1 for experiments quantifying bromide release. LB400 and RHA1 were provided 10mM biphenyl as carbon and energy source. RR1 and CB1190 are not able to grow on biphenyl and consequently RR1 was grown with 20 mM pyruvate while CB1190 was provided 5 mM of 1,4-dioxane every three days for twelve days. Cultures were grown at 30°C and shaken at 200 rpm in 160 mL bottles sealed with teflon-lined Mininert caps (VICI Valco Instruments, Houston, TX) containing 50 mL of medium. Experiments were conducted when cells had reached mid-exponential growth phase with absorbance between 0.26 and 0.27 when measured at 600 nm. Due to the propensity of CB1190 to form cell clumps and to float on the surface of the medium, cell density could not be measured spectrophotometrically. Protein measurements were taken by extracting 1 mL of sample and using the Coomassie blue protocol provided with the reagents by Thermo Scientific (Waltham, MA). Protein values averaged 26 mg/L when the experiments were started. All experiments were repeated at least twice.
Experimental Design
Our experimental methods were modified from previous studies performed to analyze PBDE transformation under anaerobic conditions (7,9). Neat mono- BDE 3 was purchased from Sigma Aldrich (St. Louis, MO); all other individual congeners were purchased from Accustandard (New Haven, CT). The mixture DE-71 was purchased from Wellington Labs (Guelph, Ontario, Canada). Vials for the PBDE transformation experiments were prepared by adding 1 μL of a stock containing 50 μg/mL PBDE congener dissolved in nonane to 8mL culture vials that were subsequently left in a laminar hood for 5 minutes to evaporate the nonane. 3 mL of bacterial culture at an optical density of 0.26–0.27 at 600 nm were added to the vials and sealed with Teflon- lined screw caps. Any observable biphenyl solids in the culture bottle were allowed to settle to the bottom before transfer of the bacteria since biphenyl solids affect extraction efficiency of the PBDEs. The final PBDE concentration in sample vials was 17 ng/mL. Controls consisted of autoclaved cells and PBDEs. All sample vials and controls were performed in triplicate. All samples were incubated in a shaking incubator at 30°C. The experimental vials were extracted after three days using 3 mL of isooctane. 0.6 ng of 2,2′,4,5′-tetrabromobiphenyl dissolved in methanol was added as an internal standard. The vials were briefly vortexed and then placed on a shaker table for 16 hours.
The bromide formation experiments were carried out by adding 4 μL of a nonane stock containing 50 mg/mL mono-BDE 3 or 1 mg/mL tetra-BDE 47 to 60 mL glass bottles with a glass syringe. The bottles were left uncapped in a laminar hood to evaporate the nonane for 5 minutes. 12 mL of cells at an optical density in the range of 0.26 – 0.27 were added to the bottles and capped with a Teflon lid. The final concentrations of mono- BDE 3 were 17 μg/mL and for tetra-BDE 47 were 3.3 μg/mL. The bottles were placed in a shaking incubator at 30°C and monitored for 24 hours. Periodically, 200 μL of sample were removed with a plastic disposable syringe, filtered with a 0.2 μm filter, and placed in centrifuge tubes kept at 4°C until the end of the experiment for bromide analysis. After 24 hours the remaining liquid in the bottle was extracted with a 1:1 volume of isooctane for PBDE analysis using 2,2′,4,5′-tetrabromobiphenyl as an internal standard. The percent transformation was determined by comparing the average concentrations of PBDEs in the triplicate live samples with the triplicate autoclaved controls. To determine if the average concentrations of samples and controls were statistically different, a student t-test was used with a p-value of 0.1. Total error was used to determine error bars.
Analytical Techniques
1mL of isooctane extract was transferred into a 2mL vial with a disposable glass pipette and analyzed by gas chromatography with electron capture detection (GC-ECD) model 3800 from Varian Inc. (Walnut Creek, CA). A 30 m DB-1 column with 0.25 mm I.D. and 0.25 μm film thickness was used. The GC temperature program was as follows: 110°C for 2 min, ramp at 30°C/min to 200°C and then ramp at 1.5°C/min until 300°C. The injector temperature was 250°C and the detector was kept at 325°C. Automatic splitless injection was used with 2 μL injection volume; flow of the nitrogen carrier gas was maintained at a constant pressure of 25 psi. Three point calibration curves, ranging from 0.5 to 50 ng/mL were run daily for all individual congeners to adjust for variation in ECD response. Extraction efficiencies, measured as the amount of PBDE recovered compared to the amount of PBDEs added to the control samples, for the di- through hexa-BDEs ranged from 73 to 118%. The extraction efficiency for mono-BDE was much less, around 54%. Extraction efficiency for the internal standard averaged 93%. Identification of hydroxylated intermediates was performed using a Waters (Milford, MA) GC-MS in electron impact mode with a 30 m DB-5 column with 0.25 mm I.D. and 0.25 μm film thickness. A splitless injection of 2 μL was made and the helium carrier gas flow was 2 mL/min. The temperature program was the same as described above. To assure good recoveries of acidic metabolites, the samples were acidified to pH4 with H2SO4 before extraction. Some of the sample aliquots were derivatized with heptafluorobutyric acid anhydride (HFBA) (Supelco, Bellefonte, PA) for improved detection. Briefly, the 3 mL samples were extracted with 1 mL of ethyl acetate by shaking overnight. The ethyl acetate was then transferred to a new vial and evaporated to dryness with a stream of nitrogen. 200 μL of acetonitrile and 50 μL of HFBA were added and the vial was placed in a 55°C oven for 90 minutes. After incubation, the acetonitrile was evaporated to dryness and the contents of the vial were resuspended in 100 μL of isooctane.
Determination of Bromide
The relatively low concentrations of bromide released during PBDE transformation and complex matrix precluded the use of conventional bromide measurement techniques. To measure bromide at low concentrations in the presence of organic matter a new sensitive analytical technique was developed in which bromide was converted to hypobromous acid (HOBr), which subsequently was detected by quantification of brominated phenols formed after reaction with phenol. This method is similar to methods by Mishra et al. (21) and Reddy-Noone et al. (22) which could not be used in our experiments due to interference from the presence of cells. A bromide calibration curve was created using standards of NaBr in deionized organic-free water. The samples were buffered with 50 μL of a 1 M KH2PO4 solution adjusted to pH7. 250 ng of phenol (10 μL from a 25 ng/μL phenol solution) and 5 μg of NaOCl (10 μL from a 0.5 μg/μL NaOCl solution) were then added. The samples were briefly vortexed and allowed to react in the dark for 10 minutes. The reaction was stopped by adding 50 μL of a 0.3 M Na2S2O3 solution. The samples were extracted with 100 μL of isooctane by vortexing for 5 minutes prior to analysis by a Varian 3800 GC-ECD with a 30 m DB-1 column with 0.25 mm I.D. and 0.25 μm film thickness with automatic splitless injection The following temperature program was used: 80°C for 2 min, then ramp at 20°C/min until 300°C and hold for 2 min. As shown in Figure SI-S1, six products are formed in this method: 2,4,6,- trichlorophenol, 2-bromo-4,6-dichlorophenol, 4-bromo-2,6-dichlorophenol, 2,6-dibromo- 4-chlorophenol, 2,4-dibromo-6-chlorophenol and 2,4,6-tribromophenol. The formation of 2,4,6-trichlorophenol and 2,4,6-tribromophenol was verified with standards from Sigma- Aldrich. Standards for the intermediate partially brominated and partially chlorinated phenols were not commercially available, so their formation was verified by GC-MS. In the absence of bromide, 2,4,6-trichlorophenol was the predominant product with small amount of a second peak (A) likely attributable to trace amounts of bromide in the NaOCl stock (Figure SI-S2). As the concentration of bromide increased, the areas of three peaks (2,4,6-tribromophenol and peaks A and B) increased as the area of 2,4,6- trichlorophenol decreased. A calibration curve in deionized water, in which the sum of the peak areas of peak A, B and 2,4,6-tribromophenol were used was linear with a modest background signal attributable to bromide in the NaOCl stock solution (Figures SI-S3 and SI-S4). To create a calibration curve with cells grown in minimal medium, the samples were filtered with a 0.2 μm filter. The filtrate was diluted 1:100 in deionized organic-free water and then spiked with bromide. A linear calibration curve was obtained for the diluted filtrate with a larger intercept related to interference from the bacterial medium and cellular material (Figure SI-S3). The yield of this method could not be ascertained directly, as standards for three of the brominated and chlorinated phenols are not commercially available. However, using 2,4,6-trichlorophenol (TCP) as a surrogate, the yield could be determined comparing the amount of TCP produced to the theoretical amount that should have been formed given the NaOCl and phenol concentrations, using rate constants from Acero et al. (23). The method was determined to be 98% efficient in nanopure water. When the reaction was repeated in the presence of 1/100 diluted filtered cells spiked with bromide, the efficiency of 2,4,6-trichlorophenol formation dropped to 60%. The limit of detection was 2.5 nM bromide in nanopure water. Over the course of an experiment, samples were collected and filtered with a 0.2 μm filter. The filtrate was then diluted 1:100 into a final volume of 2.5 mL of deionized organic-free water to minimize interference from bacterial protein in the medium. After measuring the concentrations of bromide in the diluted samples, the values were multiplied by 100 to determine the true concentration of bromide in the experimental samples. For each experiment, a new bromide calibration curve was created using filtered cells collected at the start of the experiment and covering the range of bromide expected based on PBDE transformation.
Results
Four bacterial isolates were exposed to 13 PBDE congeners ranging from mono- to hexa- BDEs for three days under aerobic conditions. The four strains were: two PCB degrading bacteria, Rhodococcus jostii RHA1 and Burkholderia xenovorans LB400, a related strain known to degrade aromatics, Rhodococcus sp. RR1 (19), and an additional ether- degrading bacterium, Pseudonocardia dioxanivorans CB1190 (24). RHA1 and LB400 were initially grown on biphenyl while RR1 and CB1190 were grown on pyruvate and 1,4-dioxane respectively.
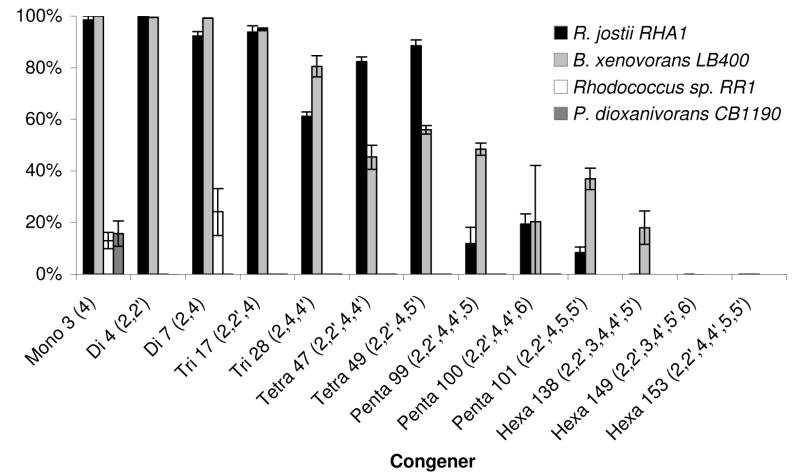
Figure 1 shows the percent transformation for the thirteen tested congeners for each of the four tested isolates. The percent of PBDE transformed was determined by comparing the PBDE concentrations remaining in the live samples with those present in the autoclaved controls in order to account for extraction efficiencies and any potential mass losses. The two PCB-degrading strains, RHA1 and LB400, transformed greater than 90% of the mono- and di-BDE congeners within three days, while they transformed only 10% to 45% of the penta-BDE congeners. Hexa-BDEs were found to be the most resistant to transformation by these strains. Of the three tested congeners, LB400 was only able to transform 18% of hexa-BDE 138 but not hexa-congeners 153 and 149, while RHA1 was not able to degrade any of the three tested congeners even at increased cell densities of 0.7 (OD600). Rhodococcus sp. strain RR1 transformed the mono and di-BDE 7 (2,4- dibromodiphenyl ether), whereas it was unable to transform other congeners including di- BDE 4 (2,2′-dibromodiphenyl ether). CB1190 transformed only about 16% of the mono- BDE and none of the more brominated congeners.
Figure 1.
Percent transformation of ten PBDE congeners by R.jostii RHA1, B. xenovorans LB400, Rhodococcus sp. RR1 and P. dioxanivorans CB1190. Values are averages of triplicates. The percent degradation was calculated as the difference between the average PBDE concentration in the controls versus the live samples. Error bars represent the total error. Strains RR1 and CB1190 were not tested for the ability to degrade hexa-BDEs.
RHA1 was tested for the ability to transform a combination of PBDE congeners by exposure to the DE-71 industrial penta-BDE mixture. RHA1 transformed 95% of the tetra-BDE 47, 78% of the penta-BDE 99 and 45% of the penta-BDE 100. Although the exact concentrations cannot be compared with those from the individual PBDE experiments due to differing PBDE and cell concentrations, the same general transformation pattern is evident, indicating that the transformation ability of RHA1 is not significantly affected by the presence of multiple PBDEs.
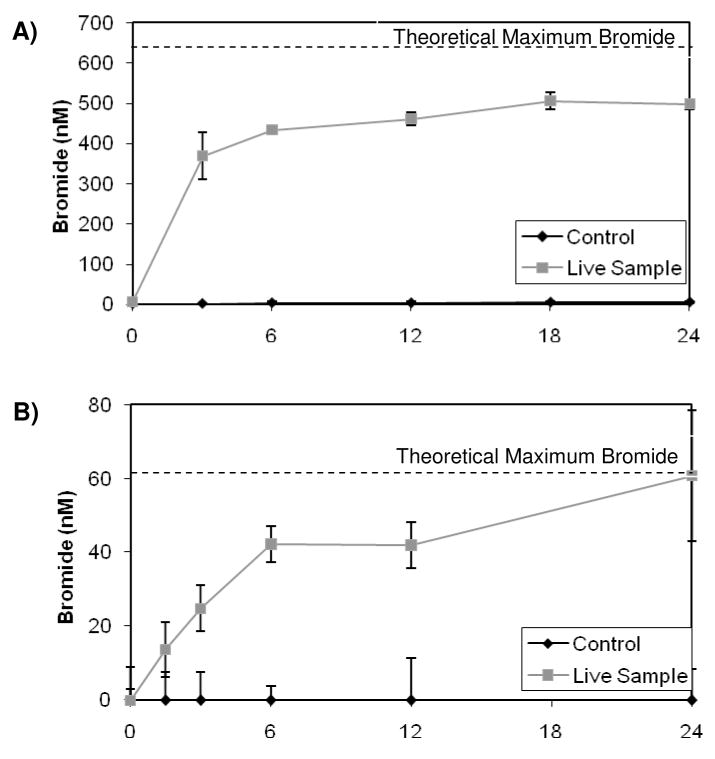
Strain RHA1 was observed to release stoichiometric quantities of chloride during the transformation of 2,2′-dichlorobiphenyl (17). LB400 has also been shown to dechlorinate certain PCBs by hydroxylation of the chlorinated carbon in the ortho position (25,26). Therefore, we analyzed for bromide generation concurrent with PBDE disappearance in order to confirm PBDE transformation by RHA1 and LB400. Since bromide detection is analytically challenging at low concentrations, mono-BDE 3 was added to RHA1 and LB400 bacterial cells at 68 μM, one thousand-fold higher than previously tested concentrations, and bromide release was monitored over a 24-hour period. The same experiment was repeated using tetra-BDE 47 at a concentration of 7.6 μM. Because of the elevated PBDE concentrations, the PBDEs were not transformed to the same extent as in the lower concentration experiments. RHA1 and LB400 transformed 99% and 66% respectively of the 68 μM mono-BDE 3 they were exposed to, whereas RHA1 only degraded 20% of the 7.6 μM tetra-BDE 47 it was exposed to and LB400 did not degrade the elevated concentrations of the tetra sufficiently to detect production of bromide. Figure 2 shows the formation of bromide by RHA1 and LB400. Figure SI-S5 shows chromatograms from the bromide analysis for the RHA1 samples and controls exposed to mono-BDE at zero and 18 hours. No significant formation of bromide was detected in the autoclaved controls. Concurrent with the degradation of mono-BDE and tetra-BDE by RHA1, 80% and 100% respectively, of the stoichiometric bromide concentrations were recovered, indicating that RHA1 is capable of cleaving the all the carbon-bromine bonds under aerobic conditions. Conversely, when LB400 transformed mono-BDE 3, only 10% of the stoichiometric quantities of bromide were recovered, indicating that LB400 transforms mono-BDE 3 to brominated intermediates rather than completely debrominating it. Since the bromide analytical method relies upon converting bromide to brominated phenols, any bromophenols generated as byproducts of PBDE transformation could skew the bromide quantification. However, no production of 4-bromophenol or 2,4-dibromophenol was detected in RHA1 samples transforming 4- bromodiphenyl ether and 2,2′,4,4′-tetrabromodiphneyl ether respectively. Nor were any bromophenols detected as by-products in the LB400 samples.
Figure 2.
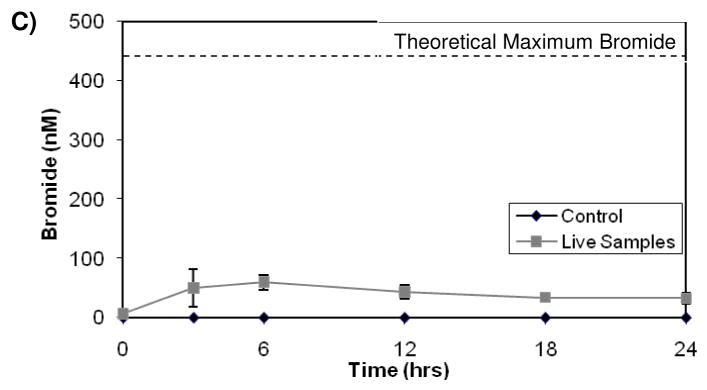
Bromide production by A) RHA1 exposed to mono-BDE 3, B) RHA1 exposed to tetra-BDE 47 and C) LB400 exposed to mono-BDE 3 over the course of 24 hours. The values are the averages for triplicates and the error bars are standard deviations. The dotted line indicates the maximum amount of bromide that could theoretically be released from the amount of PBDEs transformed.
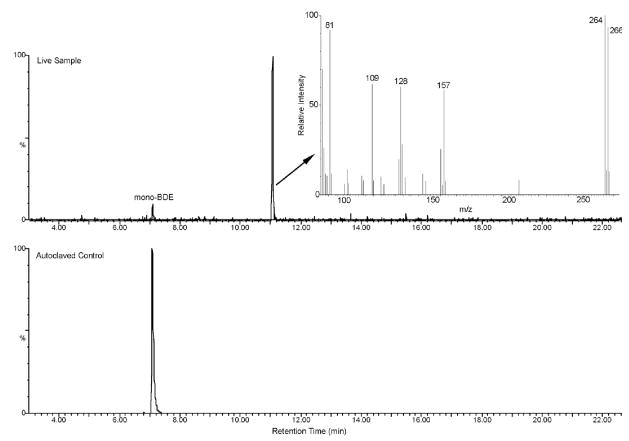
During aerobic PCB degradation by these strains, one of the rings is initially hydroxylated at two adjacent carbon positions to form a catechol which is then dehydrogenated to a dihydroxylated PCB (27). The chromatograms generated by the GC- ECD during LB400 exposure to mono-BDE contained a peak with a longer retention time than the original mono-BDE in all three of the live samples. This peak was not present in the autoclaved controls. The peak area of this unidentified compound was approximately as large as the substrate peak in the autoclaved controls. Analysis by GC-MS of acidified aliquots from these samples and comparison of the spectrum with that of a known standard identified this compound to be a hydroxylated mono-BDE. The M+ ions were 264 m/z and 266 m/z and were present at the same ratio as expected for brominated compounds containing equal proportions of the 79 and 81 bromine isotopes (Figure 3). The exact position of the hydroxyl group on the ring is unknown, although the MS fragments suggest that the hydroxyl group is on the non-brominated ring. No other brominated compounds, including dihydroxylated mono-BDE, were detected by GC-MS. Aliquots from the LB400 samples exposed to mono-BDE 3 were also derivatized with HFBA for further verification (Figure SI-S6). The spectrum yielded M+ ions of 460 m/z and 462 m/z which correspond to the molecular weight of a hydroxylated mono-BDE with the addition of HFBA. No other fluorinated peaks were detected. These results indicate that a hydroxylated mono-BDE was the dominant product of the 90% of mono- BDE transformed by LB400.
Figure 3.
GC-MS Chromatograms of underivatized extracts of an autoclaved control and a live sample of LB400 degrading 68 μM mono-BDE 3 after 24 hours. The insert is the MS spectrum of the peak at 11 minutes identified as a hydroxylated mono-BDE.
Samples of RHA1 transforming 68 μM mono-BDE 3 were analyzed for the presence of metabolites. Aliquots were acidified to pH 4 and analyzed by GC-ECD and GC-MS, but no peaks were detected that were not present in the acidified autoclaved control.
Discussion
Experiments exposing four bacterial strains to 13 mono- thorough hexa-BDE congeners over the course of three days found that PBDEs can be aerobically transformed. The aerobic PCB-degrading species Rhodococcus jostii RHA1 and Burkholderia xenovorans LB400 were found to be capable PDBE transformers, active toward some fraction of all of the mono- through penta-BDE congeners. In addition, LB400 was able to transform 18% of hexa-BDE 138, although none of the other tested hexa-BDEs were transformed by either strain. Aerobic biotransformation of PBDEs by bacteria had been previously described only for mono- through tri-BDE congeners by Sphingomonas strains (10,11,13,27). This is the first report of aerobic transformation of the environmentally relevant tetra- and penta-BDE congeners as well as one hexa-BDE. RHA1 and LB400 catalyzed much greater removal of PBDEs than the previously described Sphingomonas sp. strain PH-07. Kim et al. observed at most 20% degradation of mono-BDE 3 and di- BDEs 7 and 15 over the course of eight days and only trace removal of tri-BDE 28. By comparison, RHA1 and LB400 transformed all of the mono-BDE 3 and di-BDE 7 and transformed between 60 and 80% of tri-BDE 28 within three days.
The other two species tested in this study, Rhodococcus sp. RR1 and Pseudonocardia dioxanivorans CB1190, were unable to transform most of the tested PBDEs. Although strain RR1 degrades aromatics, it only partially transformed mono-BDE 3 and one of the di-BDEs. It appears that RR1 can only transform PBDEs with one non-brominated ring, such as mono-BDE 3 and di-BDE 7 (2,4-dibromodiphenyl ether), whereas all of the congeners that were not transformed have both rings brominated. Although CB1190 is an ether-degrading bacterium, capable of breaking the ether bond in 1,4-dioxane and tetrahydrofuran, it was able to transform only the mono-BDE 3 and only to a small extent (17%). Clearly, although they are ethers, PBDEs have very different chemical structures from the known ether substrates of CB1190. Furthermore, CB1190 has not been observed to degrade other halogenated compounds (24).
In general, transformation of the mono to hexa-BDE congeners by these bacteria was inversely proportional to the degree of bromination. Plotting the logarithm of the octanol- water partition coefficients obtained from Braekevelt et al. (28) for various PBDEs against the transformation ability of RHA1 and LB400 gives a correlation co-efficient of 0.85. The inverse correlation between biotransformation and degree of PBDE bromination was expected due to the increased size and hydrophobicity of more brominated congeners, reducing availability to the cell. Additionally, increased bromination also decreases the susceptibility of the carbons to hydroxylation and can sterically hinder enzymatic attack. This trend has also been observed with PCB transformation by RHA1 and LB400 (15–17). RHA1, however, is able to transform PCBs more effectively than the brominated diphenyl ether counterparts. For example, whereas RHA1 transformed about 82% of tetra-BDE 47 and 12% of the penta 99 within a three day incubation, it transformed on average 95% of tetra-CB 47 and 90% of penta-CB 99 in three days when tested at comparable cell densities and concentrations. Furthermore, RHA1 is able to transform hexa- and hepta-CBs, whereas no hexa-BDE transformation was observed for this strain even at higher cell densities. Although comparisons with LB400 studies are difficult because the cell densities and durations of the experiments are significantly different, Bopp found that LB400 readily transformed 59% of hexa-CB 153, whereas BDE 153 was not transformed in this study (15). This is perhaps in part due to the order of magnitude greater hydrophobicity (log Kow of 7.9 for hexa-BDE versus 6.8 for hexa-CB) and the larger molecular volume of the PBDEs compared to the PCBs, greater steric hindrance of bromide compared to chloride, as well as other factors (28).
It is difficult to determine the effect of bromine position on the transformation extent in this study due to the small variability observed among the different congeners in the same homologue group. Tri-BDE 28 (2,4,4′-tri-BDE) was transformed to a lesser extent than tri-BDE 17 (2,2′,4-tri-BDE) and tetra-BDE 49 (2,2′,4,5′-tetra-BDE) was transformed slightly more than tetra-BDE 47 (2,2′,4,4′-tetra-BDE). In the case of hexa-BDEs the effect of bromine position is more evident since only one congener was transformed. LB400 transformed 18% of 138 (2,2′,3,4,4′,5′-hexa-BDE) but not the other two congeners 153 (2,2′,4,4′,5,5′-hexa-BDE) and 149 (2,2′,3,4′,5′,6-hexa-BDE). Hexa-BDE 138 is the only tested congener that has two adjacent non-brominated carbons at the ortho and meta position where ring hydroxylation of PCBs by LB400 typically begins (29,30).
Experiments in which RHA1 and LB400 were exposed to elevated concentrations of mono-BDE 3 and tetra-BDE 47 found that bromide is released during PBDE transformation. Between 80 and 100% of the stoichiometric quantities of bromide were released when RHA1 transformed PBDEs. Because this occurred with both a mono-BDE and a tetra-BDE, which has bromines on both rings, this indicates that RHA1 completely debrominated the starting molecule. PCB dechlorination has been observed with RHA1 where stoichiometric concentrations of chloride were released from 2,2′-di-CB (17). Strain LB400 has also been shown to dechlorinate ortho-substituted PCBs (25,26). This is the first report of complete aerobic debromination of PBDEs with stoichiometric bromide. Previously tested PBDE-transforming bacteria formed a variety of products. Schmidt et al. found that Sphingomonas strain SS3 released 35% of the transformed 4-bromodiphenyl ether as bromide (11) while generating 4-bromophenol and 4-bromocatachol as byproducts. Kim et al. showed that Sphingomonas strain PH-07 converted 4-bromodiphenyl ether to 4-bromophenol and 2-hydroxymuconic acid (13).
Results with LB400 were significantly different as only 10% of the transformed mono-BDE was released as bromide with the remaining substrate transformed to a hydroxylated mono-BDE as identified by GC-MS. Previous studies with PCBs showed that LB400 could dechlorinate certain PCBs by initial hydroxylation at the chlorinated carbon (31). In this work, however, LB400 hydroxylated the mono-BDE on the non-brominated ring, suggesting that bromide is being released during subsequent transformation of the hydroxylated metabolite. Why the majority of the substrate is only transformed to the hydroxylated form is unclear, however, the release of approximately 10% bromide suggests that there may be some product toxicity or that the steps beyond the hydroxylation reaction are kinetically rate-limiting. Given that the metabolite 2,3-hydroxybiphenyl from biphenyl degradation by LB400 is known to be toxic to this strain (32), it is possible that the hydroxylated mono-BDE is toxic to the cell as well. The chlorobenzoates formed during breakdown of PCBs by LB400 are also toxic (33). The generation of hydroxylated PBDEs by this strain is potentially problematic to higher organisms as hydroxylated PBDEs are more endocrine-disrupting than their non-hydroxylated counterparts (34–36). Interestingly, LB400 might use a different enzyme to transform PBDEs than PCBs. A biphenyl dioxygenase enzyme (Bph) catalyzes the first step of PCB transformation in LB400 by adding two hydroxyl groups to the ring (37–39). With the accumulation of mono-hydroxylated mono-BDE by LB400, it appears instead that monooxygenase activity may be involved in PBDE transformation by this strain.
Aerobic PCB transformation is typically cometabolic, as has been shown with RHA1 and LB400, although certain species have been found that can metabolize 4-chlorobiphenyl (40). Sphingomonas sp. SS3 has been shown to grow on 4-bromodiphenyl ether (10,11). The ability of RHA1 and PB400 to grown on PBDEs should be investigated, as metabolic growth on PBDEs would eliminate the necessity for a primary substrate such as biphenyl.
The results of this study demonstrate that PBDEs with six bromines or less can be aerobically transformed by PCB-degrading bacteria. Depending on the species, we found that PBDEs can be transformed and release stoichiometric quantities of bromide or lead to an accumulation of hydroxylated derivatives with increased endocrine disruption characteristics. This has potential implications for the environment, as PBDE transformation could occur naturally, although further studies are required to assess the potential for these reactions to occur under environmental conditions.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Jorge Loyo Resales for assistance with GC-MS analysis and Eva Agus for help with analytical techniques. We also thank Dr. Bill Mohn and Dr. Rebecca Parales for kindly donating Rhodococcus jostii RHA1 and Pseudonocardia dioxanivorans CB1190, respectively. We gratefully acknowledge Accustandard for providing the GC-MS spectrum of a hydroxylated mono-BDE. K.R. would also like to thank Dr. Gregory Cost for helpful advice. Funding was provided by The UC Riverside Center for Water Resources, the NIEHS Superfund Basic Research Program ES04705-19 and the Chang-Lin Tien Scholarship for Biodiversity.
Footnotes
Supporting Information
Supporting information includes a detailed description of the bromide analytical method and associated figures as well as the chromatograms of the derivatized hydroxylated mono-BDE.
References
- 1.Wit d. An overview of brominated flame retardants in the environment. Chemosphere. 2002;46:583–674. doi: 10.1016/s0045-6535(01)00225-9. [DOI] [PubMed] [Google Scholar]
- 2.Meerts IATM, van Zanden JJ, Luijks EAC, van Leeuwen-Bol I, Marsh G, Jakobsson E, Bergman A, Brouwer A. Potent competitive interactions of some brominated flame retardants and related compounds with human transthyretin in vitro. Toxicol Sci. 2000;56(1):95–104. doi: 10.1093/toxsci/56.1.95. [DOI] [PubMed] [Google Scholar]
- 3.She JW, Holden A, Sharp M, Tanner M, Williams-Derry C, Hooper K. Polybrominated diphenyl ethers (PBDEs) and polychlorinated biphenyls (PCBs) in breast milk from the Pacific Northwest. Chemosphere. 2007;67(9):S307–S317. doi: 10.1016/j.chemosphere.2006.05.154. [DOI] [PubMed] [Google Scholar]
- 4.Xia K, Luo MB, Lusk C, Armbrust K, Skinner L, Sloan R. Polybrominated diphenyl ethers (PBDEs) in biota representing different trophic levels of the Hudson River, New York: From 1999 to 2005. Environ Sci Technol. 2008;42(12):4331–4337. doi: 10.1021/es703049g. [DOI] [PubMed] [Google Scholar]
- 5.Oros DR, Hoover D, Rodigari F, Crane D, Sericano J. Levels and distribution of polybrominated diphenyl ethers in water, surface sediments, and bivalves from the San Francisco Estuary. Environ Sci Technol. 2005;39(1):33–41. doi: 10.1021/es048905q. [DOI] [PubMed] [Google Scholar]
- 6.Norén K, Meironyté D. Certain organochlorine and organobromine contaminants in Swedish human milk in perspective of past 20–30 years. Chemosphere. 2000;40(9–11):1111–1123. doi: 10.1016/s0045-6535(99)00360-4. [DOI] [PubMed] [Google Scholar]
- 7.He J, Robrock KR, Alvarez-Cohen L. Microbial reductive debromination of polybrominated diphenyl ethers (PBDEs) Environ Sci Technol. 2006;40(14):4429–4434. doi: 10.1021/es052508d. [DOI] [PubMed] [Google Scholar]
- 8.Gerecke AC, Hartmann PC, Heeb NV, Kohler HP, Giger W, Schmid P, Zennegg M, Kohler M. Anaerobic degradation of decabromodiphenyl ether. Environ Sci Technol. 2005;39(4):1078–1083. doi: 10.1021/es048634j. [DOI] [PubMed] [Google Scholar]
- 9.Robrock KR, Korytar P, Alvarez-Cohen L. Pathways for the anaerobic microbial debromination of polybrominated diphenyl ethers. Environ Sci Technol. 2008;42(8):2845–2852. doi: 10.1021/es0720917. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt S, Fortnagel P, Wittich RM. Biodegradation and transformation of 4,4′-dihalodiphenyl and 2,4-dihalodiphenyl ethers by Sphingomonas Sp strain Ss33. Appl Environ Microbiol. 1993;59(11):3931–3933. doi: 10.1128/aem.59.11.3931-3933.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt S, Wittich RM, Erdmann D, Wilkes H, Francke W, Fortnagel P. Biodegradation of diphenyl ether and its monohalogenated derivatives by Sphingomonas Sp strain Ss3. Appl Environ Microbiol. 1992;58(9):2744–2750. doi: 10.1128/aem.58.9.2744-2750.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hundt K, Jonas U, Hammer E, Schauer F. Transformation of diphenyl ether by Trametes versicolor and characterization of ring cleavage products. Biodegradation. 1999;10:279–286. [Google Scholar]
- 13.Kim Y-M, Nam I-H, Murugesan K, Schmidt S, Crowley DE, Chang Y-S. Biodegradation of diphenyl ether and transformation of selected brominated congeners by Sphingomonas sp PH-07. Appl Microbio Biotechnol. 2007;77:187–194. doi: 10.1007/s00253-007-1129-z. [DOI] [PubMed] [Google Scholar]
- 14.Arnett CM, Parales JV, Haddock JD. Influence of chlorine substituents on rates of oxidation of chlorinated biphenyls by the biphenyl dioxygenase of Burkholderia sp strain LB400. Appl Environ Microbiol. 2000;66(7):2928–2933. doi: 10.1128/aem.66.7.2928-2933.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bopp L. Degradation of highly chlorinated PCBs by Pseudmonas strain LB400. Journal of Industrial Microbiology. 1986;1:23–29. [Google Scholar]
- 16.Maltseva OV, Tsoi TV, Quensen JF, Fukuda M, Tiedje JM. Degradation of anaerobic reductive dechlorination products of Aroclor 1242 by four aerobic bacteria. Biodegradation. 1999;10(5):363–371. doi: 10.1023/a:1008319306757. [DOI] [PubMed] [Google Scholar]
- 17.Seto M, Kimbara K, Shimura M, Hatta T, Fukuda M, Yano K. A novel transformation of polychlorinated biphenyls by Rhodococcus sp strain RHA1. Appl Environ Microbiol. 1995;61(9):3353–3358. doi: 10.1128/aem.61.9.3353-3358.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLeod MP, Warren RL, Hsiao WWL, Araki N, Myhre M, Fernandes C, Miyazawa D, Wong W, Lillquist AL, Wang D, Dosanjh M, Hara H, Petrescu A, Morin RD, Yang G, Stott JM, Schein JE, Shin H, Smailus D, Siddiqui AS, Marra MA, Jones SJM, Holt R, Brinkman FSL, Miyauchi K, Fukuda M, Davies JE, Mohn WW, Eltis LD. The complete genome of Rhodococcus sp RHA1 provides insights into a catabolic powerhouse. P Natl Acad Sci. 2006;103(42):15582–15587. doi: 10.1073/pnas.0607048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deeb RA, Alvarez-Cohen L. Temperature effects and substrate interactions during the aerobic biotransformation of BTEX mixtures by toluene-enriched consortia and Rhodococcus rhodochrous. Biotechnol Bioeng. 1999;62(5):526–536. [PubMed] [Google Scholar]
- 20.Sharp JO, Wood TK, Alvarez-Cohen L. Aerobic biodegradation of n-nitrosodimethylamine (NDMA) by axenic bacterial strains. Biotechnol Bioeng. 2005;89(5):608–618. doi: 10.1002/bit.20405. [DOI] [PubMed] [Google Scholar]
- 21.Mishra S, Singh V, Jain A, Verma KK. Determination of iodide by derivatization to 4-iodo-N,N-dimethylaniline and gas chromatography-mass spectrometry. Analyst. 2000;125(3):459–464. doi: 10.1039/a908363d. [DOI] [PubMed] [Google Scholar]
- 22.Reddy-Noone K, Jain A, Verma KK. Liquid-phase microextraction-gas chromatography-mass spectrometry for the determination of bromate, iodate, bromide and iodide in high-chloride matrix. J Chromatogr A. 2007;1148(2):145–151. doi: 10.1016/j.chroma.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 23.Acero JL, Piriou P, von Gunten U. Kinetics and mechanisms of formation of bromophenols during drinking water chlorination: Assessment of taste and odor development. Water Res. 2005;39(13):2979–2993. doi: 10.1016/j.watres.2005.04.055. [DOI] [PubMed] [Google Scholar]
- 24.Mahendra S, Alvarez-Cohen L. Pseudonocardia dioxanivorans sp. nov., a novel actinomycete that grows on 1,4-dioxane. Int J Sys Evol Microbiol. 2005;55(Pt 2):593–598. doi: 10.1099/ijs.0.63085-0. [DOI] [PubMed] [Google Scholar]
- 25.Seeger M, Zielinski M, Timmis KN, Hofer B. Regiospecificty of dioxygenation of di-to pentachlorobiphenyls and their degradation to chlorobenzoates by the bph-encoded catabolic pathway of Burkholderia sp. strain LB400. Appl Environ Microbiol. 1999;65(8):3614–3621. doi: 10.1128/aem.65.8.3614-3621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haddock JD, Horton JR, Gibson DT. Dihydroxylation and dechlorination of chlorinated bipenyls by purified biphenyl 2,3-dioxygenase from Pseudomonas sp. strain LB400. J Bacteriol. 1995;177(1):20–26. doi: 10.1128/jb.177.1.20-26.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bedard DL, Haberl ML. Influence of chlorine substitution pattern on the degradation of polychlorinated biphenyls by 8 bacterial strains. Microbial Ecol. 1990;20(2):87–102. doi: 10.1007/BF02543870. [DOI] [PubMed] [Google Scholar]
- 28.Braekevelt E, Tittlemier SA, Tomy GT. Direct measurement of octanol-water partition coefficients of some environmentally relevant brominated diphenyl ether congeners. Chemosphere. 2003;51(7):563–567. doi: 10.1016/S0045-6535(02)00841-X. [DOI] [PubMed] [Google Scholar]
- 29.Ahmed M, Focht DD. Degradation of polychlorinated biphenyls by 2 species of Achromobacter. Can J Microbiol. 1973;19(1):47–52. doi: 10.1139/m73-007. [DOI] [PubMed] [Google Scholar]
- 30.Furukawa K, Tomikuza N, Kamibayashi A. Effect of chlorine substitution on the bacterial metabolism of various polychlorinated biphenyls. Appl Environ Microbiol. 1979;38:301–310. doi: 10.1128/aem.38.2.301-310.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seeger M, Timmis KN, Hofer B. Degradation of chlorobiphenyls catalyzed by the bph-encoded biphenyl-2,3-dioxygenase and biphenyl-2,3-dihydrodiol-2,3-dehydrogenase of Pseudomonas sp LB400. Fems Microbiol Lett. 1995;133(3):259–264. doi: 10.1111/j.1574-6968.1995.tb07894.x. [DOI] [PubMed] [Google Scholar]
- 32.Cámara B, Herrera C, González M, Couve E, Hofer B, Seeger M. From PCBs to highly toxic metabolites by the biphenyl pathway. Environ Microbiol. 2004;6(8):842–850. doi: 10.1111/j.1462-2920.2004.00630.x. [DOI] [PubMed] [Google Scholar]
- 33.Martinez P, Agullo L, Hernandez M, Seeger M. Chlorobenzoate inhibits growth and induces stress proteins in the PCB-degrading bacterium Burkholderia xenovorans LB400. Arch Microbiol. 2007;188(3):289–297. doi: 10.1007/s00203-007-0247-4. [DOI] [PubMed] [Google Scholar]
- 34.Canton RF, Sanderson JT, Letcher RJ, Bergman A, van den Berg M. Inhibition and induction of aromatase (CYP19) activity by brominated flame retardants in H295R human adrenocortical carcinoma cells. Toxicol Sci. 2005;88(2):447–455. doi: 10.1093/toxsci/kfi325. [DOI] [PubMed] [Google Scholar]
- 35.Dingemans MML, de Groot A, van Kleef RGDM, Bergman A, van den Berg M, Vijverberg HPM, Westerink RHS. Hydroxylation increases the neurotoxic potential of BDE-47 to affect exocytosis and calcium homeostatis in PC 12 cells. Environ Health Persp. 2008;116(5):637–643. doi: 10.1289/ehp.11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meerts IATM, Letcher RJ, Hoving S, Marsh G, Bergman A, Lemmen JG, van der Burg B, Brouwer A. In vitro estrogenicity of polybrominated diphenyl ethers, hydroxylated PBDEs, and polybrominated bisphenol A compounds. Environ Health Persp. 2001;109(4):399–407. doi: 10.1289/ehp.01109399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mondello FJ. Cloning and expression in Escherichia coli of Pseudomonas strain LB400 genes encoding polychlorinated biphenyl degradation. J Bacteriol. 1989;171(3):1725–1732. doi: 10.1128/jb.171.3.1725-1732.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mondello FJ, Turcich MP, Lobos JH, Erickson BD. Identification and modification of biphenyl dioxygenase sequences that determine the specificity of polychlorinated biphenyl degradation. Appl Environ Microbiol. 1997;63(8):3096–3103. doi: 10.1128/aem.63.8.3096-3103.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hofer B, Eltis LD, Dowling DN, Timmis KN. Genetic-analysis of a Pseudomonas locus encoding a pathway for biphenyl polychlorinated biphenyl degradation. Gene. 1993;130(1):47–55. doi: 10.1016/0378-1119(93)90345-4. [DOI] [PubMed] [Google Scholar]
- 40.Field JA, Sierra-Alvarez R. Microbial transformation and degradation of polychlorinated biphenyls. Environ Poll. 2008;155(1):1–12. doi: 10.1016/j.envpol.2007.10.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.