Abstract
Intracellular bacterial pathogens have adapted their metabolism to optimally utilize the nutrients available in infected host cells. We recently reported the identification of an asparagine transporter required specifically for cytosolic multiplication of Francisella. In the present work, we characterized a new member of the major super family (MSF) of transporters, involved in isoleucine uptake. We show that this transporter (here designated IleP) plays a critical role in intracellular metabolic adaptation of Francisella. Inactivation of IleP severely impaired intracellular F. tularensis subsp. novicida multiplication in all cell types tested and reduced bacterial virulence in the mouse model. To further establish the importance of the ileP gene in F. tularensis pathogenesis, we constructed a chromosomal deletion mutant of ileP (ΔFTL_1803) in the F. tularensis subsp. holarctica live vaccine strain (LVS). Inactivation of IleP in the F. tularensis LVS provoked comparable intracellular growth defects, confirming the critical role of this transporter in isoleucine uptake. The data presented establish, for the first time, the importance of isoleucine utilization for efficient phagosomal escape and cytosolic multiplication of Francisella and suggest that virulent F. tularensis subspecies have lost their branched-chain amino acid biosynthetic pathways and rely exclusively on dedicated uptake systems. This loss of function is likely to reflect an evolution toward a predominantly intracellular life style of the pathogen. Amino acid transporters should be thus considered major players in the adaptation of intracellular pathogens.
INTRODUCTION
Francisella tularensis is a Gram-negative bacterium causing the zoonotic disease tularemia in a broad variety of animal species (1). Humans can be infected by this highly infectious pathogen by different means, including direct contact with sick animals, inhalation, insect bites, or ingestion of contaminated water or food. Four different subspecies (subsp.) of F. tularensis that differ in virulence and geographic distribution exist, designated subsp. tularensis, holarctica, mediasiatica, and novicida. The four major subspecies of F. tularensis cause a fulminant disease in mice that is similar to tularemia in humans (2). Although F. tularensis subsp. novicida is rarely pathogenic in humans, its genome shares approximately 97% nucleotide sequence identity with the human-pathogenic species (3), suggesting that homologous proteins function via similar mechanisms. F. tularensis subsp. novicida is thus widely used as a model to study highly virulent subspecies (4).
Francisella virulence is tightly linked to its capacity to multiply intracellularly in the cytosolic compartment of infected host cells (5, 6). Macrophages are one of the primary intracellular niches for Francisella survival in the host. Recent studies from our group (7–9) and from several others groups (10–13) have highlighted the importance of nutrient acquisition and metabolic adaptations in Francisella pathogenesis. In particular, amino acids are thought to represent major sources of carbon and energy for F. tularensis (13–15). However, the mammalian cell cytosol contains variable amounts of amino acids. In particular, cells are auxotrophic for a number of amino acids, including Leu, Ile, Val, and Thr (16), and must therefore rely mainly on their import from the sources available in the external medium.
Interestingly, a recent study has shown that F. tularensis was able to boost the intracellular levels of all amino acids by triggering the host macroautophagy degradation machinery (13). To utilize these intracellular sources of amino acids, Francisella is equipped with a battery of transporters, including numerous secondary transporters (15). Several other intracellular pathogens have recently been shown to require amino acids as important carbon, energy, and nitrogen sources (17–20). For example, the two intravacuolar pathogens Anaplasma phagocytophilum and Legionella pneumophila have evolved with efficient strategies to boost the levels of host amino acids (21).
In Legionella pneumophila, the protein PhtA, a member of the subclass of the major facilitator superfamily of transporters designated phagosomal transporters (Pht), is required for threonine uptake in the Legionella-containing vacuole (22). The L. pneumophila genome encodes 10 additional PhtA paralogues, some of which are also required during intracellular replication (23). PhtJ is required for acquisition of valine (24), whereas the roles of the phtE and phtF loci in the L. pneumophila genome are currently unknown. Interestingly, PhtC and PhtD were very recently shown to contribute to protecting L. pneumophila from dTMP starvation (25). Thus, Pht proteins in Legionella contribute not only to the assimilation of amino acids but also to additional metabolic functions.
Phylogenetic studies revealed that PhtA orthologues were encoded by a variety of other bacterial genomes but exclusively among intracellular pathogenic bacteria (24, 26). We have recently characterized the first member of the Pht family in Francisella, the asparagine transporter AnsP (9). Our study revealed that AnsP was required for asparagine assimilation by intracellular Francisella when the bacteria were present in the cytosolic compartment of infected cells, indicating that the functions of the Pht transporters are not restricted to bacteria residing exclusively in a phagosomal compartment.
In the present study, we addressed the role of a second member of the Pht family highly conserved in all the Francisella genomes, the protein FTN_1654 in F. tularensis subsp. novicida. We chose to focus on this putative amino acid transporter protein because the FTN_1654 gene has been repeatedly found in earlier genetic screens to be required for replication of F. tularensis subsp. novicida in vivo (11, 27, 28). Moreover, its orthologue in F. tularensis subsp. tularensis Schu S4 (FTT_0056c) has been shown to be required for normal bacterial replication in the hepatocytic human cell line HepG2 (29). Furthermore, inactivation of FTL_1803, the orthologue of FTN_1654 in the F. tularensis subsp. holarctica live vaccine strain (LVS), also resulted in altered replication in HepG2 human hepatic cells and reduced virulence in the mouse (30). Transport function mediated by FTN_1654 thus appears to be important in vitro as well as in vivo and is not compensated by any other putative transporter encoded by the Francisella genome.
We show here that the protein FTN_1654 is an isoleucine transporter (hence, designated IleP). IleP-mediated isoleucine uptake appears to be vital for bacterial intracellular multiplication and virulence in both F. tularensis subsp. novicida and F. tularensis subsp. holarctica LVS. Remarkably, genome comparisons suggest that specialization toward an intracellular lifestyle has led pathogenic Francisella subspecies to lose their branched-chain amino acid (BCAA) pathways to rely exclusively on transporter-mediated acquisition of these amino acids from the host.
MATERIALS AND METHODS
Ethics statement.
All experimental procedures involving animals were conducted in accordance with guidelines established by the French and European regulations for the care and use of laboratory animals (decrees 87-848, 2001-464, 2001-486, and 2001-131 and European Directive 2010/63/UE) and approved by the INSERM Ethics Committee (authorization number 75-906).
Bacterial strains and plasmids.
F. tularensis subsp. novicida strain U112, the F. tularensis subsp. holarctica LVS (kindly provided by A. Sjostedt), and their derivatives were grown in the following liquid media: Schaedler broth (bioMérieux, Marcy l'Etoile, France), tryptic soy broth (TSB) supplemented with cysteine (Becton, Dickinson and Co.), and Chamberlain chemically defined medium (CDM) (31). Bacteria were grown at 37°C on either of the following solid media: premade chocolate agar (PolyViteX; bioMérieux SA, Marcy l'Etoile, France) or chocolate plates prepared from GC medium base, IsoVitalex vitamins, and hemoglobin (BD Biosciences, San Jose, CA, USA).
All bacterial strains, plasmids, and primers used in this study are listed in Table S1 in the supplemental material.
Bioinformatic analyses.
Secondary structure predictions were performed using the programs HMM (available at http://www.cbs.dtu.dk/services/TMHMM-2.0/) and TMRPres2D for visual representation of transmembrane protein models (available at http://bioinformatics.biol.uoa.gr/TMRPres2D).
Construction of chromosomal deletion mutants.
We inactivated the ileP gene in the wild-type (WT) F. tularensis subsp. novicida strain U112 (FTN_1654) and in the wild-type F. tularensis LVS by allelic replacement, resulting in the deletion of the entire gene (from the start codon to the stop codon). For this, we used the counterselectable plasmid pMP812 (32). The recombinant plasmid pMP812-ΔFTN_1654 was constructed by overlap PCR. Primers p1 and p2 amplified the 1,046-bp region upstream of position +1 of the FTN_1654 coding sequence, and primers p2 and p3 amplified the 1,089-bp region immediately downstream of the FTN_1654 stop codon (see Table S1 in the supplemental material). Primers p2/p3 have an overlapping sequence of 20 nucleotides, resulting in complete deletion of the FTN_1654 coding sequence after crossover PCR. PCRs with primers p1/p2 and p3/p4 were performed with exTaq polymerase (Fermentas). The products were purified using a QIAquick PCR purification kit (QIAgen, CA). One hundred nanograms of each amplification product was used as a template for PCR with primers p1/p4 and treated with 30 cycles of PCR (94°C for 30 s, 54°C for 30 s, and 72°C for 120 s). The gel-purified 2,135-bp fragment was digested with BamHI and NotI (New England BioLabs) and cloned into BamHI-NotI-digested pMP812 (32). The plasmid was introduced into F. tularensis subsp. novicida U112 by electroporation. A standard two-step allelic exchange procedure was used to create the chromosomal ΔFTN_1654 mutant (14). The mutant strains were checked for loss of the corresponding wild-type genes by PCR sequencing (GATC Biotech) using specific primers.
We also generated a chromosomal deletion of the gene orthologous to F. tularensis subsp. novicida ileP in the F. tularensis subsp. holarctica LVS (FTL_1803). We used the recombinant plasmid pMP812-ΔFTN_1654 (previously used to create the ΔFTN_1654 deletion). The plasmid was introduced into F. tularensis LVS by cryo-transformation. The same two-step allelic exchange procedure was used to create the chromosomal ΔFTL_1803 mutant (14).
Functional complementation.
The plasmid used for complementation of the F. tularensis subsp. novicida ΔileP mutant (ΔFTN_1654), pKK214-pgro-FTN_1654, was constructed by overlap PCR. Primers p9/p10 amplified the wild-type FTN_1654 gene (1,305 bp), and the primers p7/p8 amplified 333 bp of the pgro promoter. Primers p8/p10 have an overlapping sequence of 20 nucleotides. PCRs with the p9/p10 and p7/p8 primer pairs were performed with exTaq polymerase (Fermentas), and the products were purified using a QIAquick PCR purification kit (QIAgen, CA). A 200 μM concentration of each amplification product was used as a template for PCR with primers p11/p13. The gel-purified 1,638-bp fragment was digested with SmaI and PstI (New England BioLabs) and cloned into SmaI-PstI-digested pKK214. The plasmids pKK214 (empty plasmid) and pKK214-pgro-FTN_1654 (complementing plasmid) were introduced into U112 and the F. tularensis subsp. novicida ΔileP mutant (ΔFTN_1654) by electroporation. The same plasmid was used for complementation of the F. tularensis LVS ΔileP mutant (ΔFTL_1803).
Growth kinetics in broth.
Stationary-phase bacterial cultures of wild-type F. tularensis subsp. novicida U112 or LVS and the U112 ΔileP or LVS ΔileP mutant strains were diluted at a final optical density at 600 nm (OD600) of 0.1 in tryptic soy broth (TSB) or CDM. The OD600 of the culture was measured every hour during an 8-h period. F. tularensis LVS and its isogenic mutant were grown in standard Chamberlain chemically defined medium (CDM) while F. tularensis subsp. novicida and its isogenic mutant were grown in the optimized CDM devoid of threonine and valine (CDMmin) (9).
Isolation of total RNA.
Bacteria were centrifuged for 2 min in a microcentrifuge at room temperature, and the pellet was quickly resuspended in TRIzol solution (Invitrogen, Carlsbad, CA). Samples were either processed immediately or frozen and stored at 80°C. Samples were treated with chloroform, and the aqueous phase was used in an RNeasy Cleanup protocol (Qiagen, Valencia, CA) with an on-column DNase digestion of 30 min.
Quantitative real-time RT-PCR.
F. tularensis U112 and the U112 ΔileP mutant strain were grown at 37°C to an OD600 of 0.1 in Schaedler broth containing vitamin K3 (Schaedler-K3 broth). After 4 h of incubation, samples were harvested, and RNA was isolated. One microgram of RNA was reverse transcribed using random hexamers and Superscript II reverse transcriptase (Invitrogen) according to the protocol provided by the manufacturer. Real-time reverse transcription-PCR (RT-PCR) was performed with gene-specific primers using an ABI Prism 7700 system and SYBR green PCR master mix (Applied Biosystems, Foster City, CA). To calculate the amount of gene-specific transcript, a standard curve was plotted for each primer set using a series of diluted genomic DNAs from strain U112. The amounts of FTN_1654, FTN_1653, and FTN_1655 transcripts were normalized to the helicase gene (FTN_1594) transcript, whose expression is only slightly changed during growth.
Transport assays.
Bacteria were grown in Chamberlain medium to mid-exponential phase and then harvested by centrifugation and washed twice with Chamberlain medium without amino acids. Bacteria were suspended at a final OD600 of 0.5 in the same medium containing 50 mg ml−1 of chloramphenicol. After 15 min of preincubation at 25°C, uptake was started by the addition of either l-[U-14C]isoleucine or l-[U-14C]leucine (Ile and Leu; PerkinElmer) at various concentrations (ranging from 1 to 100 μM). The 14C-radiolabeled amino acids were at a specific activity of 7.4 GBq mmol−1. Samples (100 μl of bacterial suspension) were removed at regular intervals and collected by vacuum filtration on membrane filters (type HA, 25 mm, 0.22-μm pore size; Millipore) and rapidly washed with Chamberlain medium without amino acids (2 times with 5 ml). Filters were transferred to scintillation vials and counted in a Hidex 300 scintillation counter. The counts per minute (cpm) were converted to picomoles of amino acid taken up per sample using a standard derived by counting a known quantity of the same isotope under similar conditions.
Multiplication in macrophages.
J774.1 (ATCC TIB-67), THP-1 (ATCC TIB-202), and HepG2 (ATCC HB 8065) cells were propagated in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum. Cells were seeded at a concentration of 2 × 105 cells per well in 12-well cell tissue plates, and monolayers were used at 24 h after seeding. J774.1, THP-1, and HepG2 monolayers were incubated for 60 min at 37°C with the bacterial suspensions (multiplicity of infection of 100 for J774.1 and THP-1 and of 1,000 for HepG2) to allow the bacteria to enter. After a washing step (time zero of the kinetic analysis), the cells were incubated in fresh culture medium containing gentamicin (10 μg ml−1) to kill extracellular bacteria.
At several time points, cells were washed three times in DMEM, macrophages were lysed by addition of water, and the titer of viable bacteria released from the cells was determined by spreading preparations on chocolate agar plates. For each strain and time in an experiment, the assay was performed in triplicate. Each experiment was independently repeated at least three times, and the data presented originate from one typical experiment. For suppression experiments with the ΔileP strain, amino acids were added to 3 mM at the time of infection and maintained throughout the infection.
Confocal experiments.
Colocalization experiments were performed in J774-1 cells. With F. tularensis subsp. novicida, macrophages were grown in DMEM containing 8 μM threonine, and with F. tularensis LVS, macrophages were grown in DMEM containing 80 μM isoleucine. Assays were performed as described previously (33). Cells were examined using a ×63 oil immersion objective on a Leica TSP SP5 confocal microscope. Colocalization tests were performed by using ImageJ software, and mean numbers were calculated on more than 500 cells for each condition. Confocal microscopy analyses were performed at the Cell Imaging Facility (Faculté de Médecine Necker Enfants-Malades).
Mouse infections.
Wild-type F. tularensis subsp. novicida and mutant strains were grown in TSB to exponential growth phase and diluted to the appropriate concentrations. Six- to 8-week-old female BALB/c mice (Janvier, Le Genest St. Isle, France) were intraperitoneally (i.p.) inoculated with 200 μl of bacterial suspension. For supplementation experiments with the ΔileP strain, threonine was injected i.p. three times into mice (50 μl of a 5 mM solution per mouse at 14 h before infection, upon infection, and 24 h after infection).
The actual number of viable bacteria in the inoculum was determined by plating dilutions of the bacterial suspension on chocolate plates. After 2 days, mice were sacrificed. Homogenized spleen and liver tissue from each mouse was diluted and spread onto chocolate agar plates.
Intracellular amino acid quantification assays.
Ten million THP-1 cells in RPMI medium were infected by the F. tularensis LVS for 1 h as previously described (34). After cells were washed with RPMI medium containing 10 μg/ml gentamicin, they were further incubated at 37°C for 24 h. Cells were then pelleted and immediately frozen in liquid nitrogen. They were then submitted to amino acid quantification.
Amino acid detection and quantification were done according to a standard Agilent procedure using a Zorbax Eclipse-AAA high-performance liquid chromatography (HPLC) column. Briefly, the samples and the amino acid standard solutions were automatically derivatized with ortho-phthalaldehyde (OPA) by programming the robotic auto sampler. After derivatization, an amount equivalent to 10 or 20 μl of each sample was injected on a Zorbax Eclipse-AAA column (5-μm pore size; 150 by 4.6 mm [Agilent]) at 40°C with fluorescence detection. The aqueous mobile phase was 40 mM NaH2PO4, adjusted to pH 7.8 with NaOH, while the organic mobile phase was acetonitrile-methanol-water (45:45:10, vol/vol/vol). The separation was obtained at a flow rate of 2 ml min−1 with a gradient program that allowed for 2 min at 0% B followed by a 16-min step that raised eluent B (acetonitrile-methanol-water; 45:45:10, vol/vol/vol) to 60%. Then washing at 100% B and equilibration at 0% B were performed in a total analysis time of 38 min.
RESULTS
IleP, a new member of the major facilitator superfamily (MFS) of transporters.
The region of ileP (FTN_1654 in F. tularensis subsp. novicida) is highly conserved in all F. tularensis subspecies (Fig. 1A). The IleP protein is also highly conserved and shares 99.3% identity with its orthologues in F. tularensis subsp. holarctica (FTL_1803) and F. tularensis subsp. tularensis (FTT_0056c) (see Fig. S1 in the supplemental material). Like most MFS members, IleP is predicted to comprise 12 transmembrane helixes (Fig. 1B). The closest homologue in other bacterial species is LLO_3121 of Legionella longbeachae (designated PhtJ) that shares 40.3% amino acid identity with the Francisella IleP proteins (see Fig. S1). The conserved amino acids between the Francisella and L. longbeachae proteins are globally scattered along the protein sequence. Of note, the valine transporter PhtJ of L. pneumophila (Lpg1893) shares 47.2% and 33.3% amino acid identity with the L. longbeachae protein LLO_3121 and FTN_1654, respectively
FIG 1.
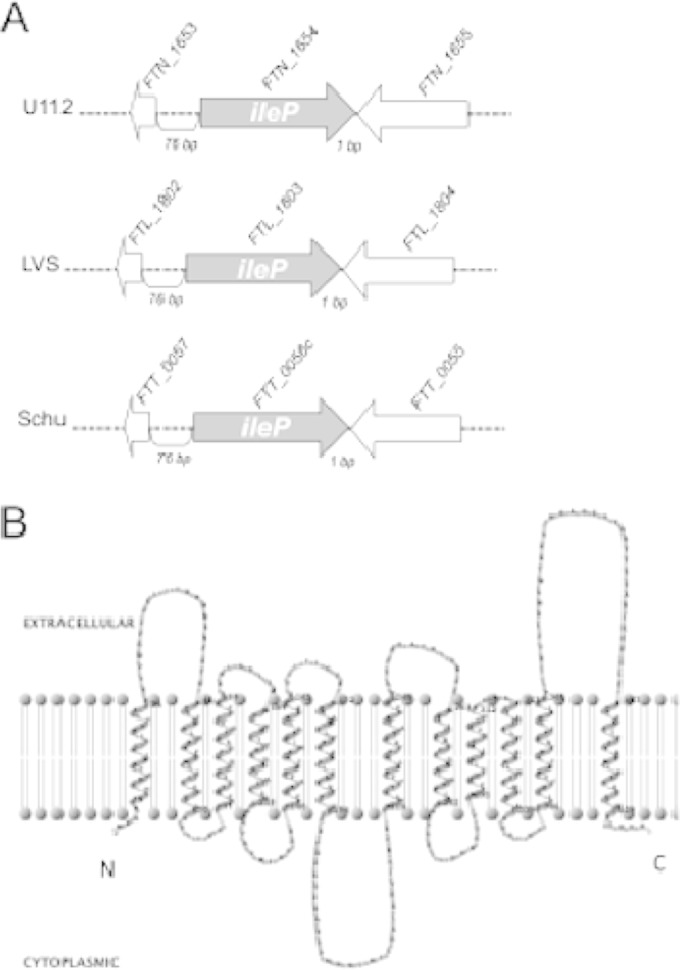
The transporter IleP. (A) The gene ileP (FTN_1654) of F. tularensis subsp. novicida (U112) is flanked by two genes in opposite orientation (FTN_1653, upstream, and FTN_1655, downstream), separated by short intergenic regions (76 bp and 1 bp, respectively). FTN_1654 shares 98% identity with its orthologues in F. tularensis subsp. holarctica (FTL_1803) and F. tularensis subsp. tularensis (FTT_0056c). This region is also highly conserved in F. tularensis subsp. holarctica LVS (LVS) and the F. tularensis subsp. tularensis Schu S4 (Schu) strain. (B) The IleP protein of F. tularensis subsp. novicida shares 99.3% identity with its orthologues in F. tularensis subsp. holarctica (FTL_1803) and F. tularensis subsp. tularensis (FTT_0056c). The protein IleP is predicted to comprise 12 transmembrane helixes, with the N- and C-terminal extremities facing the cytoplasm. Two-dimensional modeling of Ile folding was performed using the HMM program and TMRPres2D for visual representation.
The ileP gene of F. tularensis subsp. novicida is predicted to constitute a single transcriptional unit, according to the BioCyc collection of pathway/genome databases. We generated a chromosomal deletion of the ileP gene in F. tularensis subsp. novicida U112 by allelic replacement (see Materials and Methods). The deletion had no polar effect on the expression of the two flanking genes (see Fig. S2 in the supplemental material).
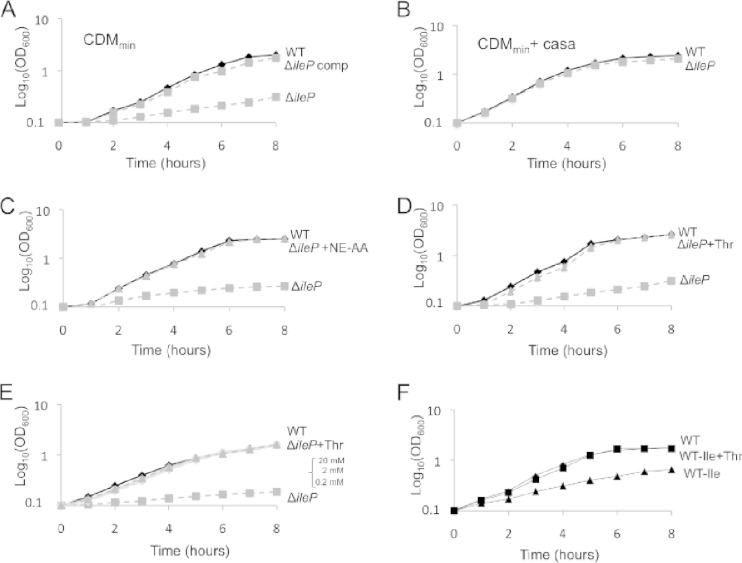
We first evaluated the impact of the ΔileP deletion on bacterial growth in a chemically defined medium optimized for F. tularensis subsp. novicida (CDMmin), described in Gesbert et al. (9). The ΔileP mutant showed a severe growth defect in this medium, and wild-type growth was restored in the ΔileP complemented strain (Fig. 2A). Supplementation of the CDMmin with 0.5% Casamino Acids (Fig. 2B) restored multiplication of the ΔileP mutant to the wild-type level, suggesting that the growth defect of the ΔileP mutant in nonsupplemented CDMmin was due to impaired amino acid transport. Supplementation of the CDMmin with a 10-fold excess of either a pool of essential amino acids (arginine, histidine, lysine, methionine, tyrosine, and cysteine) or useful but nonessential amino acids (isoleucine, leucine, aspartic acid, serine, and proline) failed to alleviate the growth defect of the ΔileP mutant (data not shown). In contrast, CDMmin supplementation with a pool of the nine nonessential amino acids (threonine, valine, alanine, glutamate, glutamine, glycine, phenylalanine, tryptophan, and asparagine) restored wild-type growth to the ΔileP mutant (Fig. 2C). Therefore, we next supplemented the CDMmin individually with each of the nonessential amino acids. Threonine was the only amino acid that restored growth of the ΔileP mutant to the wild-type level (Fig. 2D) at all the concentrations tested (Fig. 2E).
FIG 2.
Growth in chemically defined medium (CDM). (A) Growth of wild-type F. tularensis subsp. novicida, the ΔileP strain, and the ΔileP complemented strain (ΔileP-comp) was assayed in the optimized CDM medium for F. tularensis subsp. novicida as described in Gesbert et al. (9). This medium contains 11 amino acids: arginine, lysine, histidine, tyrosine, methionine, cysteine, isoleucine, leucine, aspartic acid, serine, and proline. WT, wild-type F. tularensis subsp. novicida; WT pKK(−), WT carrying pKK214; ΔileP, WT with a deletion of the ileP gene; ΔileP pKK(−), ΔileP strain carrying pKK214; ΔileP-comp, ΔileP strain carrying pKK214-pgro-ileP. (B) CDMmin supplemented with Casamino Acids (casa; final concentration, 0.5%). (C) CDMmin with or without the addition of an equimolar mixture (3 mM final) of nonessential amino acids (NE AA; threonine, valine, alanine, glutamate, glutamine, glycine, phenylalanine, tryptophan, and asparagine). (D) CDMmin with (+Thr) or without threonine supplementation (final concentration, 3 mM). (E) CDMmin with (+Thr) or without supplementation with decreasing concentrations of threonine (final concentration, 0.2 mM to 20 mM). (F) Wild-type F. tularensis subsp. novicida was grown in CDMmin (WT) or CDMmin lacking isoleucine, with (WT −Ile +Thr) or without (WT −Ile) threonine supplementation (final concentration, 3 mM).
Since threonine is nonessential for growth, we then reasoned that threonine could be used by the bacterium for the biogenesis of the amino acid(s) that could not be taken up by the ΔileP mutant. We therefore replaced individually each of the 11 amino acids present in the CDMmin by threonine and monitored growth of the wild-type strain. Isoleucine was the only amino acid that could be replaced by threonine without altering the normal growth of F. tularensis subsp. novicida (Fig. 2F). Of note, F. tularensis subsp. novicida is equipped with an intact BCAA biosynthetic pathway (see Fig. S3 in the supplemental material) that uses threonine as precursor for BCAAs, including isoleucine.
Interestingly, wild-type F. tularensis subsp. novicida in CDMmin devoid of isoleucine was much less affected in growth than the ΔileP mutant in normal CDM (see Fig. S4A in the supplemental material), suggesting that IleP might transport an additional substrate present in the CDMmin. Furthermore, the fact that growth of the ΔileP mutant is similarly affected in CDMmin, with or without leucine (see Fig. S4A), supports the notion that IleP is not a major leucine transporter of F. tularensis subsp. novicida.
To confirm that IleP was indeed involved in isoleucine transport, we compared the uptake of radiolabeled isoleucine ([14C]Ile) by wild-type F. tularensis subsp. novicida to that of the ΔileP mutant (Fig. 3A). At isoleucine concentrations ranging from 2.5 to 25 μM, [14C]Ile incorporation was significantly affected in the ΔileP mutant (approximately 50% incorporation relative to the wild type). The fact that isoleucine transport was not totally abolished in the ΔileP mutant suggests that isoleucine could still enter via other transporter(s) in this strain. Isoleucine belongs to the group of branched-chain amino acids (BCAAs) that also comprises leucine and valine. Since isoleucine and leucine are both required for growth of Francisella in CDMmin, we decided to measure the uptake of radiolabeled leucine in wild-type F. tularensis subsp. novicida and in the ΔileP mutant (Fig. 3B). The uptake of [14C]Leu was not affected at 2.5, 5, and 10 μM leucine in the ΔileP mutant. Reduced uptake (approximately 25% reduction compared to wild type) was observed only at the highest leucine concentration (25 μM). Wild-type uptake was restored in the complemented strain with both substrates. Together, these assays revealed that IleP is mainly involved in isoleucine transport.
FIG 3.
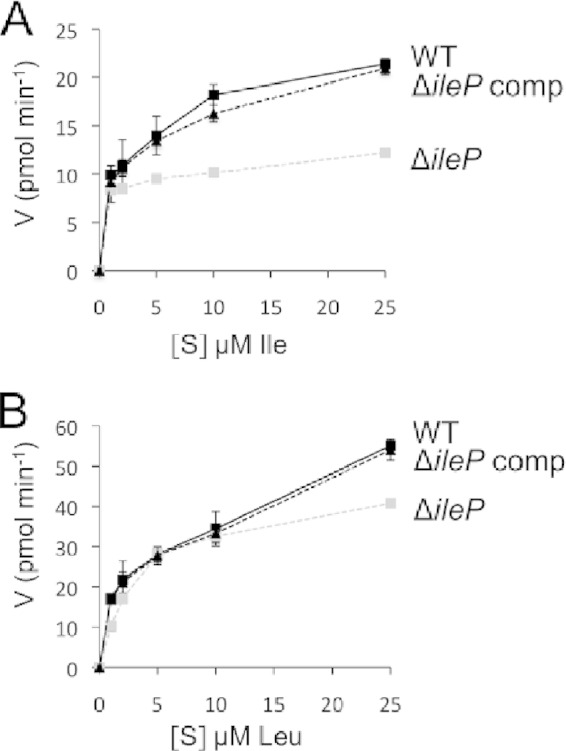
Transport assays. Kinetics of uptake of 14C-radiolabeled isoleucine (A) or leucine (B) by wild-type F. tularensis subsp. novicida (WT), the ΔileP mutant, and the ΔileP complemented strain (ΔileP comp) at substrate (S) concentrations ranging from 1 to 100 μM. V, velocity of uptake.
IleP is required for intracellular multiplication and virulence.
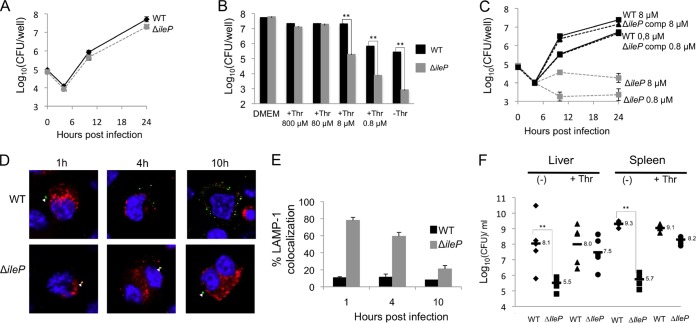
We first examined the ability of the F. tularensis subsp. novicida wild-type and ΔileP strains to survive and multiply in murine macrophage-like J774-1 cells (Fig. 4). In standard DMEM, the intracellular multiplication of the ΔileP mutant was essentially identical to that of wild-type F. tularensis subsp. novicida (Fig. 4A). In contrast, in DMEM containing limiting concentrations of threonine (Fig. 4B), multiplication of the ΔileP mutant was severely impaired compared to that of the wild type. After 24 h, cells infected with the ΔileP mutant showed a 150-fold reduction of intracellular bacteria in DMEM supplemented with 8 μM or 0.8 μM threonine compared to cells infected with the wild-type strain and a 300-fold reduction in DMEM without threonine (Fig. 4B). We further carried out a kinetics of intracellular multiplication with the wild-type, ΔileP, and ΔileP complemented strains in J774.1 cells grown in DMEM supplemented with either 8 μM or 0.8 μM threonine (Fig. 4C).
FIG 4.
Impact of ΔileP inactivation on F. tularensis subsp. novicida virulence. (A and B) Intracellular multiplication of the ΔileP mutant in J774.1 murine macrophage-like cells was essentially identical to that of wild-type F. tularensis subsp. novicida (WT) in standard DMEM (A). Under limiting concentrations of threonine (using modified DMEM, containing decreasing concentrations of threonine), multiplication of the ΔileP mutant was significantly impaired compared to that of the wild type (from 8 μM and lower concentrations of threonine) (B). Intracellular replication was monitored, after 24 h of infection, by enumerating CFU on chocolate agar plates. Each experiment was performed in triplicate. **, P < 0.01 (as determined by Student's t test). (C) Kinetics of intracellular multiplication of wild-type F. tularensis subsp. novicida (WT), the ΔileP strain, and the complemented (ΔileP comp) strain in J774.1 macrophages was monitored in DMEM supplemented with either 80 μM or 0.8 μM threonine. (D and E) Confocal microscopy. J774.1 cells were incubated for 1 h with wild-type F. tularensis subsp. novicida or the ΔileP mutant strain, and their colocalization with the phagosomal marker LAMP-1 was observed by confocal microscopy. The phagosomes of J774.1 cells were labeled with anti-LAMP-1 antibody (1/100 final dilution). Cell nuclei were labeled with 4′,6-diamidino-2-phenylindole. Bacteria (white triangles) were labeled with the primary mouse monoclonal antibody anti-F. tularensis subsp. novicida (1/500 final dilution). (C) The color images represent wild-type F. tularensis subsp. novicida (WT) and ΔileP bacteria (green), phagosomes (red), and nuclei (blue). (D) Quantification of bacteria/phagosome colocalization at 1 h, 4 h, and 10 h for the WT and ΔileP strains. **, P < 0.01 (as determined by Student's t test). (F) In vivo dissemination. Groups of five female BALB/c mice were infected intraperitoneally (i.p.) with either wild-type F. tularensis subsp. novicida (WT) or ΔileP mutant strain (200 CFU per mouse). The data represent the number of bacteria per organ (spleen and liver) for each mouse at 48 h after infection. Results are shown as log10 CFU per organ ± standard deviations.**, P < 0.01 (as determined by Student's t test). −, non-pretreated mice; +Thr, mice injected i.p. three times with 50 μl of a 5 mM solution of threonine (14 h before infection, upon infection, and at 24 h after infection).
Of note, in HepG2 human hepatocytic cells, the F. tularensis subsp. novicida ΔileP mutant showed a 3-fold reduction of intracellular bacteria after 10 h and a 10-fold reduction after 24 h in standard DMEM compared to the number of wild-type bacteria at same time points (see Fig. S5 in the supplemental material). Constitutive expression of the wild-type gene ileP (ΔileP complemented strain) restored normal intracellular replication. The fact that the ΔileP mutant failed to multiply normally in HepG2 cells under threonine-replete conditions (800 μM threonine) could be due, for example, to a limiting intracellular pool of threonine in this cell line.
Subcellular localization.
We followed the subcellular localization of the ΔileP mutant in infected cells under limiting threonine concentration (8 μM) (see Materials and Methods). Intracellular localization of the bacteria or LAMP-1 (used as a specific marker of phagosomes) was analyzed using specific antibodies, and their colocalization was monitored at three time points (1 h, 4 h, and 10 h postinfection) (Fig. 4D). Quantification of each colocalization was performed with ImageJ software (Fig. 4E). In cells infected with wild-type F. tularensis subsp. novicida, colocalization of bacteria with LAMP-1 was around only 10% after 1 h and remained in the same range throughout the infection, showing that most bacteria escape the phagosome rapidly. In contrast, approximately 80% of ΔileP mutant bacteria colocalized with LAMP-1 after 1 h of infection. After 4 h, 60% colocalization was still observed, which decreased to 21% after 10 h of infection. These data suggest that, after 10 h, most of the ΔileP mutant bacteria had escaped in the host cytosol. However, after 24 h (Fig. 4B), the number of ΔileP mutant bacteria remained 1,000-fold lower than that of the wild type (i.e., approximately at the same level as that recorded at the beginning of the infection), implying that ileP inactivation affected both phagosomal escape and cytosolic multiplication.
Virulence.
To investigate the fate of the bacteria inside host tissues, groups of five female BALB/c mice were infected intraperitoneally (i.p.) with either the wild-type F. tularensis subsp. novicida or the ΔileP mutant strain (Fig. 4F), and we monitored the number of viable bacteria in the spleen and liver. The numbers of viable bacteria were determined 48 h after infection by plating diluted tissue homogenates. The number of wild-type bacteria reached approximately 108 and 109 bacteria per liver and spleen, respectively. In contrast, ΔileP mutant bacteria were detected in both the spleen and liver at significantly lower levels than in the wild-type strain (approximately 4,000-fold fewer viable mutant bacteria were recorded in the spleen, and 400-fold fewer were found in the liver). Strikingly, upon treatment of the mice with threonine (three i.p. injections with 50 μl of a 5 mM solution of threonine) (see Materials and Methods), the number of ΔileP mutant bacteria increased significantly (approximately 100-fold in liver and 300-fold in spleen), close to the values recorded with the wild-type strain. This experiment demonstrated that threonine supplementation in vivo also restored virulence of the F. tularensis subsp. novicida ΔileP mutant (Fig. 4F).
IleP of F. tularensis LVS is required for intracellular survival under isoleucine-limiting conditions.
Although F. tularensis subsp. novicida has been extensively used as a model to study the molecular bases of Francisella pathogenicity (4), discrepancies between the phenotype of mutants generated in F. tularensis subsp. novicida and in more virulent subspecies have been occasionally reported (35). The live vaccine strain (LVS) is an attenuated strain of the highly pathogenic species F. tularensis subsp. holarctica that was originally developed as a vaccine and retains 99.92% identity to its parental species (3). Both F. tularensis subsp. novicida and the LVS are approved for use in biosafety level 2 (BSL2) laboratories and are readily genetically manipulated (36).
To further establish the importance of the ileP gene to all F. tularensis subspecies, we constructed a chromosomal deletion mutant (ΔFTL_1803) in F. tularensis LVS and evaluated its impact on intracellular multiplication. We first evaluated the impact of the mutation in standard CDM (31) (Fig. 5). Growth of the LVS ΔileP mutant (ΔFTL_1803) was only slightly affected in standard CDM (containing 1.5 mM isoleucine). Remarkably, under limiting isoleucine conditions (0.15 mM), multiplication of wild-type LVS was essentially not affected, whereas that of the mutant was almost totally abolished (in spite of the presence of threonine in the medium) (Fig. 5A). Indeed, both the F. tularensis subsp. holarctica LVS and virulent F. tularensis subsp. tularensis Schu S4 possess a defective branched-chain amino acid biosynthetic pathway (Fig. 6). Hence, in contrast to F. tularensis subsp. novicida, these subspecies are most likely unable to biosynthesize branched-chain amino acids from threonine.
FIG 5.
IleP of LVS (FTL_1803) is required for growth under isoleucine-limiting conditions. (A) Growth in broth. In standard CDM (containing 1.5 mM Ile) (31), growth of the ΔileP mutant (ΔFTL_1803) was slightly affected. When the concentration of Ile was reduced to 0.15 mM, multiplication of the wild-type LVS was essentially not affected, whereas that of the mutant was almost totally abolished. (B, C, and D) Growth in cells. Intracellular multiplication of the ΔileP mutant strain (ΔFTL_1803) was monitored in J774.1 macrophages after 24 h of infection and compared to that of the wild-type LVS in DMEM containing decreasing concentrations of Ile (B). The 80 μM Ile concentration (boxed) was chosen for further analyses. Intracellular multiplication of the ΔileP mutant strain (ΔFTL_1803) was monitored in DMEM supplemented with 80 μM Ile in J774.1 (C) and THP-1 (D) macrophages and compared to that in the wild-type LVS. (E and F) J774.1 cells were incubated for 1 h with the wild-type LVS or the ΔileP mutant strain (ΔFTL_1803), and their colocalization with the phagosomal marker LAMP-1 was observed by confocal microscopy. The phagosomes of J774.1 cells were labeled with anti-LAMP-1 antibody (1/100 final dilution). Cell nuclei were labeled with 4′,6-diamidino-2-phenylindole. Bacteria (white triangles) were labeled with primary mouse monoclonal antibody anti-Francisella (1/500 final dilution). The color images represent wild-type LVS (WT) and ΔileP bacteria (green), phagosomes (red), and nuclei (blue). Quantification of bacteria/phagosome colocalization at 1 h, 4 h, and 10 h for WT and ΔileP strains is shown in the bar graph. **, P < 0.01 (as determined by Student's t test). comp, complemented strain.
FIG 6.
Isoleucine uptake and the BCAA biosynthetic pathway in Francisella subspecies. (A) Schematic representation of isoleucine (Ile) entry in F. tularensis subsp. novicida and F. tularensis LVS. Isoleucine crosses the bacterial cytoplasmic membrane via the MFS transporter IleP. In F. tularensis subsp. novicida, when threonine is available in the medium, threonine enters the bacterium (via a dedicated transporter) and can serve as a precursor for the biosynthesis of branched-chain amino acids (BCAAs) such as isoleucine, leucine, and valine. In contrast, in F. tularensis LVS, isoleucine can only be obtained from the uptake of external isoleucine sources (via IleP and possibly other nonspecific permeases). (B) Schematic representation of the isoleucine-valine operon in F. tularensis subsp. novicida U112 (U112), F. tularensis subsp. tularensis Schu S4 (Schu), and F. tularensis subsp. holarctica LVS (LVS) strains. The intact isoleucine-valine genes are shown in blue, the inactivated genes in LVS and Schu S4 genomes are in pink, the flanking genes (eng and mfd) are in gray.
We next followed the kinetics of intracellular multiplication of the wild-type F. tularensis LVS and its ΔileP derivative (Fig. 5B and C). In standard DMEM, the ΔileP mutant of LVS showed no significant defect in intracellular multiplication in J774.1 macrophages. In contrast, the ΔileP mutant showed a significant reduction of intracellular multiplication after 24 h compared to the parental F. tularensis LVS under isoleucine-limiting conditions (100-fold decrease in number of mutant bacteria relative to wild-type bacteria at 80 μM) (Fig. 5C).
The intracellular multiplication of the ΔileP mutant was also impaired in THP-1 macrophages compared to that of the wild-type F. tularensis LVS under isoleucine-limiting conditions (Fig. 5D).
Confocal microscopy analyses of the F. tularensis LVS ΔileP mutant fully supported those obtained with the F. tularensis subsp. novicida ΔileP mutant (Fig. 5E and F). Indeed, 78% of ΔileP bacteria colocalized with LAMP-1 after 1 h of infection, and 50% colocalization was still observed after 4 h. Colocalization of the wild-type F. tularensis LVS with LAMP-1 was around only 52% after 1 h and decreased to 10% throughout the infection.
These results confirmed that ileP inactivation affected both phagosomal escape and cytosolic multiplication in the two F. tularensis subspecies.
Metabolic analyses.
L. pneumophila has been shown to trigger an important entry of amino acids upon infection of human embryonic kidney cells (16). This prompted us to evaluate the impact of F. tularensis LVS infection on the intracellular pools of BCAAs (isoleucine, leucine, and valine) in THP-1 human monocytes. Comparative measurements of free amino acids in lysates of THP-1 macrophages (see Fig. S6 in the supplemental material) showed that at 1 h postinfection, wild-type LVS triggered a significant rise in the concentration of each of the BCAAs (i.e., 13-fold, 10-fold, and 17-fold for isoleucine, leucine, and valine, respectively). Remarkably, after 24 h, the concentration of all three BCAAs significantly decreased, reaching 2- to 3-fold that of noninfected cells. We used as a nonreplicating control an iglC mutant of F. tularensis LVS. The iglC gene is part of the Francisella pathogenicity island (FPI) and encodes the intracellular growth locus C protein (IglC) that is highly induced following F. tularensis infection of macrophages (21). As with most of the other FPI mutants, iglC mutants fail to grow in macrophages and are deficient in their ability to escape from phagosomes (37). The increase in intracellular BCAA concentrations recorded at 1 h was approximately half that of the wild-type strain; and after 24 h, the BCAA concentrations were only moderately reduced (notably for Ile, at 5-fold that of noninfected cells).
We also used L. monocytogenes, another intracellular pathogen able to multiply in macrophages and with a cytosolic lifestyle, as a control. We monitored the BCAA pools in THP-1 cells infected with L. monocytogenes strain EGD-e at 1 h and 5 h postinfection. Notably, EGD-e, similar to the wild-type F. tularensis LVS, triggered a significant rise in the concentration of each of the BCAAs (i.e., approximately 10-fold for isoleucine, leucine, and valine and less than 2-fold for threonine). However, the concentrations of all three BCAAs varied only very moderately after 5 h, when bacteria had undergone several rounds of multiplication.
DISCUSSION
We characterized a new MFS transporter of Francisella that is involved in isoleucine uptake (IleP) and required for bacterial virulence. Inactivation of IleP severely impaired intracellular multiplication of both F. tularensis subsp. novicida and the F. tularensis subsp. holarctica LVS in all cell types tested.
Importantly, the data presented establish for the first time the importance of isoleucine utilization for efficient phagosomal escape and cytosolic multiplication of Francisella and suggest that virulent F. tularensis subspecies have lost their branched-chain amino acid biosynthetic pathways and rely exclusively on dedicated uptake systems.
Importance of isoleucine in phagosomal egress and cytosolic multiplication.
The branched-chain amino acids (BCAAs) are among the nine essential amino acids for humans and therefore must be supplied in the diet. The BCAA concentration in human blood serum is relatively high (in the 100 μM range) (33). In contrast, the BCAA concentration in the cytosol of human cells in culture is variable and critically depends on their concentration in the growth medium (38). In host tissues, BCAA distribution is unknown but is likely to be quite variable and also dependent on available external sources.
BCAA metabolism lies at the crossroads of several other bacterial metabolic pathways. For example, BCAA and fatty acid catabolism are used for ATP and energy production by many proteobacteria (39). In Francisella, however, BCAA degradation pathways are predicted to be nonfunctional (according to the KEGG database). This suggests that BCAAs may be mainly used for protein synthesis in these species.
We have recently shown that infection of human macrophages by Francisella triggered the rapid upregulation of the neutral amino acid transporter SLC1A5 (solute carrier family 1, member 5) and the concomitant downregulation of SLC7A5 (34). This upregulation was assumed to stimulate the entry of amino acids. Here, we found that the F. tularensis LVS triggered the uptake of important amounts of BCAAs upon entry into THP-1 macrophages, further suggesting that the bacterium has evolved a strategy of host cell manipulation to obtain amino acids in sufficient amounts for its optimal intracellular multiplication. Strikingly, the intracellular BCAA concentration sharply increased after 1 h of infection, which was followed by a strong decrease after 24 h, suggesting that these amino acids have been consumed during the course of intracellular bacterial multiplication. One may imagine that this rapid supply of amino acids is particularly useful for Francisella in the phagosome to promote protein synthesis. The fact that in cells infected with the nonreplicating iglC mutant the reduction of BCAA levels between 1 h and 24 h was lower (approximately 2-fold) than observed with wild-type bacteria (more than 5-fold) further supports the notion that these amino acids have been utilized by multiplying bacteria.
Remarkably, infection by L. monocytogenes also triggered a rapid increase of the BCAA intracellular pool at 1 h, but this pool remained almost constant after several hours of intracellular bacterial multiplication, suggesting that BCAAs are not (or marginally) used by multiplying bacteria and/or that their uptake is continuously stimulated by the infection.
Napier et al. recently showed that biotin biosynthesis was required in the Francisella phagosome to promote rapid bacterial escape (12), thus providing the first example of a metabolic requirement of Francisella in this compartment.
The phagosome into which Francisella transiently resides (less than 30 min) is a very dynamic entity (40). Hence, its composition is likely to be continuously changing from its initial formation until its disruption. One can imagine that a rapid bacterial adaptation to the available metabolites present in the phagosome may play an important role in phagosomal egress, notably by promoting efficient protein synthesis (including of factors involved in phagosomal disruption).
Interestingly, a recent study from Lobel et al. (41) revealed that several biosynthetic pathways were induced during L. monocytogenes growth in macrophage cells, including the BCAA pathway, suggesting that limited amounts of isoleucine, leucine, and valine are available in the host cytosol. The authors demonstrated that the isoleucine-responsive regulator CodY was responsible for the upregulation of L. monocytogenes virulence genes under these conditions, suggesting that the low intracellular concentrations of BCAAs might serve as signal for the bacteria to sense their intracellular location. F. tularensis genomes do not encode any CodY orthologue. However, it cannot be excluded that the intracellular isoleucine concentration might influence Francisella gene expression via yet unknown sensory mechanisms.
At any rate, one should bear in mind that nutrient concentrations in the host can affect many different host functions. For example, BCAAs have been shown to positively regulate mTOR, a Ser/Thr kinase that functions as a master regulator of various types of cellular processes, including protein synthesis, through Rag GTPase signaling. mTOR activity also promotes immune function, including reactive oxygen species (ROS) and cytokine production (42). It is thus possible that the fluctuations in BCAA concentrations result from complex host responses to bacterial infection.
The BCAA pathway, a prototypic example of patho-adaptation?
Independently of their subcellular multiplication niches, intracellular bacterial pathogens generally possess smaller genomes than their related nonpathogenic relatives as a result of reductive genome evolution (43). This evolutionary process is associated with the loss of many pathways. Accordingly, the maximal genome reduction has been observed for obligate intracellular bacteria (44). Indeed, Chlamydiaceae are auxotrophic for most amino acids, cofactors, and purine and pyrimidine nucleotides (45). Consequently, they rely on the import of these host-derived compounds, but only very few transport proteins have been characterized functionally in these species. Of interest, having observed the close proximity of chlamydial inclusions and lysosomes, Ouellette et al. were able to show that Chlamydia required lysosomal proteases and lysosomal-mediated degradation of exogenous proteins for intracellular growth, even under growth conditions in which free amino acids were present in the extracellular medium (46). That study thus suggested that the degradation of cargo derived from the endocytic pathway by the lysosome provides essential nutrients for growth of Chlamydia.
A recent study, combining proteomics, mutant analyses, and computational approaches revealed that Salmonella virulence depended on the simultaneous exploitation of numerous different host nutrients, including vitamins, carbohydrates, and amino acids (47). Comparisons of the predicted nutrient utilization and biosynthetic pathways of a series of other mammalian pathogens confirmed that most pathogens shared the capability to utilize multiple nitrogen and carbon sources. Furthermore, the systematic prediction of biosynthetic pathways commonly lost during the specialization toward pathogenesis revealed that many pathogens lost the capacity to biosynthesize several amino acids, consistent with the notion that pathogenic species obtain such nutrients from the host. For example, L. pneumophila is auxotrophic for several amino acids (including Cys, Met, Arg, Thr, Val, Ile, Leu, Phe, and Tyr) (18, 48) and thus absolutely depends on efficient strategies to acquire them from the host. Remarkably, although Mycobacterium tuberculosis is prototrophic for all 20 amino acids, it still relies on two membrane transporters to capture aspartate and asparagine to exploit these amino acids as a nitrogen source during infection (19, 20). Thus, bacteria may favor amino acid uptake to biosynthesis, not only for obvious energetic reasons but also because it offers a simple and rapid means to adapt to changing environments.
BCAAs are among the most abundant amino acids in proteins, and maintaining their pools is, thus, a prerequisite for high-level synthesis of proteins (49). Fully supporting this notion, we found that the percentages of leucine and isoleucine in F. tularensis subsp. novicida proteins (9.7% and 9.4%, respectively) were also approximately two times higher than those of the other amino acids (see Table S2 in the supplemental material). Notably, 42.3% of the proteins contain ≥10% leucine, and 36.3% contain ≥10% isoleucine (data not shown).
Selective genome reduction in F. tularensis is illustrated by the fact that the genomes of the F. tularensis subsp. tularensis SCHU S4 and F. tularensis subsp. holarctica LVS contain more than 250 pseudogenes, whereas there are only 14 in the F. tularensis subsp. novicida U112 genome (3). Remarkably, loss of gene function affects metabolic pathways. Notably, regarding amino acid biogenesis, F. tularensis subsp. novicida is predicted to possess only three incomplete amino acid synthesis pathways, whereas the subspecies tularensis and holarctica have several additional incomplete pathways (15, 50).
Our genome comparisons suggest that the pathogenic F. tularensis subsp. holarctica and F. tularensis subsp. tularensis have lost the capacity to biosynthesize BCAAs (Fig. 6) and thus require efficient, dedicated uptake systems to survive the limiting isoleucine conditions they may encounter during a mammalian infection.
In conclusion, the analysis of IleP in F. tularensis subsp. novicida and the F. tularensis LVS revealed the conserved importance of isoleucine acquisition in Francisella pathogenesis. Of note, although IleP is primarily involved in isoleucine (and to a lesser extent in leucine) transport, IleP may transport another unknown substrate that may contribute to the observed phenotypes. Our study also highlighted the importance of the cell growth conditions (in particular the utilization of carbon and nitrogen sources) in studies of the function of transporters in different subspecies. It is likely that a number of other nutrients and dedicated transporters, important for Francisella intracellular survival, still need to be discovered and functionally characterized.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Sjostedt for providing the Francisella strains U112 and LVS. We thank Karin Meibom for helpful comments and suggestions on the manuscript.
These studies were supported by INSERM, CNRS, and Université Paris Descartes, Paris Cité Sorbonne. G.G. was funded by a fellowship from the Délégation Générale à l'Armement, and E.R. was supported by a fellowship from the Région Ile de France. We acknowledge the technical platform Bioprofiler for provision of HPLC chromatography facilities.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02579-14.
REFERENCES
- 1.Sjostedt A. 2011. Special topic on Francisella tularensis and tularemia. Front Microbiol 2:86. doi: 10.3389/fmicb.2011.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wayne Conlan J, Oyston PC. 2007. Vaccines against Francisella tularensis. Ann N Y Acad Sci 1105:325–350. doi: 10.1196/annals.1409.012. [DOI] [PubMed] [Google Scholar]
- 3.Rohmer L, Fong C, Abmayr S, Wasnick M, Larson Freeman TJ, Radey M, Guina T, Svensson K, Hayden HS, Jacobs M, Gallagher LA, Manoil C, Ernst RK, Drees B, Buckley D, Haugen E, Bovee D, Zhou Y, Chang J, Levy R, Lim R, Gillett W, Guenthener D, Kang A, Shaffer SA, Taylor G, Chen J, Gallis B, D'Argenio DA, Forsman M, Olson MV, Goodlett DR, Kaul R, Miller SI, Brittnacher MJ. 2007. Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human-pathogenic strains. Genome Biol 8:R102. doi: 10.1186/gb-2007-8-6-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallagher LA, Ramage E, Jacobs MA, Kaul R, Brittnacher M, Manoil C. 2007. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc Natl Acad Sci U S A 104:1009–1014. doi: 10.1073/pnas.0606713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santic M, Asare R, Skrobonja I, Jones S, Abu Kwaik Y. 2008. Acquisition of the vacuolar ATPase proton pump and phagosome acidification are essential for escape of Francisella tularensis into the macrophage cytosol. Infect Immun 76:2671–2677. doi: 10.1128/IAI.00185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong A, Celli J. 2010. The Francisella intracellular life cycle: toward molecular mechanisms of intracellular survival and proliferation. Front Microbiol 1:138. doi: 10.3389/fmicb.2010.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alkhuder K, Meibom KL, Dubail I, Dupuis M, Charbit A. 2009. Glutathione provides a source of cysteine essential for intracellular multiplication of Francisella tularensis. PLoS Pathog 5:e1000284. doi: 10.1371/journal.ppat.1000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramond E, Gesbert G, Rigard M, Dairou J, Dupuis M, Dubail I, Meibom K, Henry T, Barel M, Charbit A. 2014. Glutamate utilization couples oxidative stress defense and the tricarboxylic acid cycle in Francisella phagosomal escape. PLoS Pathog 10:e1003893. doi: 10.1371/journal.ppat.1003893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gesbert G, Ramond E, Rigard M, Frapy E, Dupuis M, Dubail I, Barel M, Henry T, Meibom K, Charbit A. 2014. Asparagine assimilation is critical for intracellular replication and dissemination of Francisella. Cell Microbiol 16:434–449. doi: 10.1111/cmi.12227. [DOI] [PubMed] [Google Scholar]
- 10.Schulert GS, McCaffrey RL, Buchan BW, Lindemann SR, Hollenback C, Jones BD, Allen L-A. 2009. Francisella tularensis genes required for inhibition of the neutrophil respiratory burst and intramacrophage growth identified by random transposon mutagenesis of strain LVS. Infect Immun 77:1324–1336. doi: 10.1128/IAI.01318-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng K, Monack DM. 2010. Indoleamine 2,3-dioxygenase 1 is a lung-specific innate immune defense mechanism that inhibits growth of Francisella tularensis tryptophan auxotrophs. Infect Immun 78:2723–2733. doi: 10.1128/IAI.00008-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Napier BA, Meyer L, Bina JE, Miller MA, Sjostedt A, Weiss DS. 2012. Link between intraphagosomal biotin and rapid phagosomal escape in Francisella. Proc Natl Acad Sci U S A 109:18084–18089. doi: 10.1073/pnas.1206411109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steele S, Brunton J, Ziehr B, Taft-Benz S, Moorman N, Kawula T. 2013. Francisella tularensis harvests nutrients derived via ATG5-independent autophagy to support intracellular growth. PLoS Pathog 9:e1003562. doi: 10.1371/journal.ppat.1003562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raghunathan A, Shin S, Daefler S. 2010. Systems approach to investigating host-pathogen interactions in infections with the biothreat agent Francisella. Constraints-based model of Francisella tularensis. BMC Syst Biol 4:118. doi: 10.1186/1752-0509-4-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meibom KL, Charbit A. 2010. Francisella tularensis metabolism and its relation to virulence. Front Microbiol 1:140. doi: 10.3389/fmicb.2010.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price CT, Al-Quadan T, Santic M, Rosenshine I, Abu Kwaik Y. 2011. Host proteasomal degradation generates amino acids essential for intracellular bacterial growth. Science 334:1553–1557. doi: 10.1126/science.1212868. [DOI] [PubMed] [Google Scholar]
- 17.Santic M, Abu Kwaik Y. 2013. Nutritional virulence of Francisella tularensis. Front Cell Infect Microbiol 3:112. doi: 10.3389/fcimb.2013.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price CT, Richards AM, Von Dwingelo JE, Samara HA, Abu Kwaik Y. 2014. Amoeba host-Legionella synchronization of amino acid auxotrophy and its role in bacterial adaptation and pathogenic evolution. Environ Microbiol 16:350–358. doi: 10.1111/1462-2920.12290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gouzy A, Larrouy-Maumus G, Bottai D, Levillain F, Dumas A, Wallach JB, Caire-Brandli I, de Chastellier C, Wu TD, Poincloux R, Brosch R, Guerquin-Kern JL, Schnappinger D, Sorio de Carvalho LP, Poquet Y, Neyrolles O. 2014. Mycobacterium tuberculosis exploits asparagine to assimilate nitrogen and resist acid stress during infection. PLoS Pathog 10:e1003928. doi: 10.1371/journal.ppat.1003928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gouzy A, Larrouy-Maumus G, Wu TD, Peixoto A, Levillain F, Lugo-Villarino G, Guerquin-Kern JL, de Carvalho LP, Poquet Y, Neyrolles O. 2013. Mycobacterium tuberculosis nitrogen assimilation and host colonization require aspartate. Nat Chem Biol 9:674–676. doi: 10.1038/nchembio.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abu Kwaik Y, Bumann D. 2013. Microbial quest for food in vivo: “nutritional virulence” as an emerging paradigm. Cell Microbiol 15:882–890. doi: 10.1111/cmi.12138. [DOI] [PubMed] [Google Scholar]
- 22.Sauer JD, Bachman MA, Swanson MS. 2005. The phagosomal transporter A couples threonine acquisition to differentiation and replication of Legionella pneumophila in macrophages. Proc Natl Acad Sci U S A 102:9924–9929. doi: 10.1073/pnas.0502767102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fonseca MV, Swanson MS. 2014. Nutrient salvaging and metabolism by the intracellular pathogen Legionella pneumophila. Front Cell Infect Microbiol 4:12. doi: 10.3389/fcimb.2014.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sauer JD. 2006. Contribution of Pht transporters to nutrient acquisition and differentiation of intracellular Legionella pneumophila. University of Michigan, Ann Arbor, MI. [Google Scholar]
- 25.Fonseca MV, Sauer JD, Crepin S, Byrne B, Swanson MS. 2014. The phtC-phtD locus equips Legionella pneumophila for thymidine salvage and replication in macrophages. Infect Immun 82:720–730. doi: 10.1128/IAI.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen DE, Podell S, Sauer JD, Swanson MS, Saier MH Jr. 2008. The phagosomal nutrient transporter (Pht) family. Microbiology 154:42–53. doi: 10.1099/mic.0.2007/010611-0. [DOI] [PubMed] [Google Scholar]
- 27.Weiss DS, Brotcke A, Henry T, Margolis JJ, Chan K, Monack DM. 2007. In vivo negative selection screen identifies genes required for Francisella virulence. Proc Natl Acad Sci U S A 104:6037–6042. doi: 10.1073/pnas.0609675104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moule MG, Monack DM, Schneider DS. 2010. Reciprocal analysis of Francisella novicida infections of a Drosophila melanogaster model reveal host-pathogen conflicts mediated by reactive oxygen and imd-regulated innate immune response. PLoS Pathog 6:e1001065. doi: 10.1371/journal.ppat.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin A, Mann BJ. 2006. Identification of transposon insertion mutants of Francisella tularensis tularensis strain Schu S4 deficient in intracellular replication in the hepatic cell line HepG2. BMC Microbiol 6:69. doi: 10.1186/1471-2180-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marohn ME, Santiago AE, Shirey KA, Lipsky M, Vogel SN, Barry EM. 2012. Members of the Francisella tularensis phagosomal transporter subfamily of major facilitator superfamily transporters are critical for pathogenesis. Infect Immun 80:2390–2401. doi: 10.1128/IAI.00144-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chamberlain RE. 1965. Evaluation of live tularemia vaccine prepared in a chemically defined medium. Appl Microbiol 13:232–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LoVullo ED, Molins-Schneekloth CR, Schweizer HP, Pavelka MS Jr. 2009. Single-copy chromosomal integration systems for Francisella tularensis. Microbiology 155:1152–1163. doi: 10.1099/mic.0.022491-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krasnova IN, Cherkas Iu V, Denisenko TV, Kartsova LA. 1999. A quantitative analysis of amino acids in blood serum by isocratic reverse-phase HPLC. Klin Lab Diagn 7:11–14. (In Russian.) [PubMed] [Google Scholar]
- 34.Barel M, Meibom K, Dubail I, Botella J, Charbit A. 2012. Francisella tularensis regulates the expression of the amino acid transporter SLC1A5 in infected THP-1 human monocytes. Cell Microbiol 14:1769–1783. doi: 10.1111/j.1462-5822.2012.01837.x. [DOI] [PubMed] [Google Scholar]
- 35.Kingry LC, Petersen JM. 2014. Comparative review of Francisella tularensis and Francisella novicida. Front Cell Infect Microbiol 4:35. doi: 10.3389/fcimb.2014.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barker JR, Klose KE. 2007. Molecular and genetic basis of pathogenesis in Francisella tularensis. Ann N Y Acad Sci 1105:138–159. doi: 10.1196/annals.1409.010. [DOI] [PubMed] [Google Scholar]
- 37.Nano FE, Schmerk C. 2007. The Francisella pathogenicity island. Ann N Y Acad Sci 1105:122–137. doi: 10.1196/annals.1409.000. [DOI] [PubMed] [Google Scholar]
- 38.Piez KA, Eagle H. 1958. The free amino acid pool of cultured human cells. J Biol Chem 231:533–545. [PubMed] [Google Scholar]
- 39.Kazakov AE, Rodionov DA, Alm E, Arkin AP, Dubchak I, Gelfand MS. 2009. Comparative genomics of regulation of fatty acid and branched-chain amino acid utilization in proteobacteria. J Bacteriol 191:52–64. doi: 10.1128/JB.01175-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clemens DL, Lee BY, Horwitz MA. 2005. Francisella tularensis enters macrophages via a novel process involving pseudopod loops. Infect Immun 73:5892–5902. doi: 10.1128/IAI.73.9.5892-5902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lobel L, Sigal N, Borovok I, Ruppin E, Herskovits AA. 2012. Integrative genomic analysis identifies isoleucine and CodY as regulators of Listeria monocytogenes virulence. PLoS Genet 8:e1002887. doi: 10.1371/journal.pgen.1002887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soliman GA. 2013. The role of mechanistic target of rapamycin (mTOR) complexes signaling in the immune responses. Nutrients 5:2231–2257. doi: 10.3390/nu5062231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bliska JB, Casadevall A. 2009. Intracellular pathogenic bacteria and fungi–a case of convergent evolution? Nat Rev Microbiol 7:165–171. doi: 10.1038/nrmicro2049. [DOI] [PubMed] [Google Scholar]
- 44.Merhej V, Georgiades K, Raoult D. 2013. Postgenomic analysis of bacterial pathogens repertoire reveals genome reduction rather than virulence factors. Brief Funct Genomics 12:291–304. doi: 10.1093/bfgp/elt015. [DOI] [PubMed] [Google Scholar]
- 45.Omsland A, Sixt BS, Horn M, Hackstadt T. 2014. Chlamydial metabolism revisited: interspecies metabolic variability and developmental stage-specific physiologic activities. FEMS Microbiol Rev 38:779–801. doi: 10.1111/1574-6976.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ouellette SP, Dorsey FC, Moshiach S, Cleveland JL, Carabeo RA. 2011. Chlamydia species-dependent differences in the growth requirement for lysosomes. PLoS One 6:e16783. doi: 10.1371/journal.pone.0016783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steeb B, Claudi B, Burton NA, Tienz P, Schmidt A, Farhan H, Maze A, Bumann D. 2013. Parallel exploitation of diverse host nutrients enhances Salmonella virulence. PLoS Pathog 9:e1003301. doi: 10.1371/journal.ppat.1003301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eylert E, Herrmann V, Jules M, Gillmaier N, Lautner M, Buchrieser C, Eisenreich W, Heuner K. 2010. Isotopologue profiling of Legionella pneumophila: role of serine and glucose as carbon substrates. J Biol Chem 285:22232–22243. doi: 10.1074/jbc.M110.128678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sonenshein AL. 2007. Control of key metabolic intersections in Bacillus subtilis. Nat Rev Microbiol 5:917–927. doi: 10.1038/nrmicro1772. [DOI] [PubMed] [Google Scholar]
- 50.Huber B, Escudero R, Busse HJ, Seibold E, Scholz HC, Anda P, Kampfer P, Splettstoesser WD. 2010. Description of Francisella hispaniensis sp. nov., isolated from human blood, reclassification of Francisella novicida (Larson et al. 1955) Olsufiev et al. 1959. as Francisella tularensis subsp. novicida comb. nov. and emended description of the genus Francisella. Int J Syst Evol Microbiol 60:1887–1896. doi: 10.1099/ijs.0.015941-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.