Abstract
The aim of this study was to elucidate the function of the plasmid-borne mcp (methyl-accepting chemotaxis protein) gene, which plays pleiotropic roles in Cronobacter sakazakii ATCC 29544. By searching for virulence factors using a random transposon insertion mutant library, we identified and sequenced a new plasmid, pCSA2, in C. sakazakii ATCC 29544. An in silico analysis of pCSA2 revealed that it included six putative open reading frames, and one of them was mcp. The mcp mutant was defective for invasion into and adhesion to epithelial cells, and the virulence of the mcp mutant was attenuated in rat pups. In addition, we demonstrated that putative MCP regulates the motility of C. sakazakii, and the expression of the flagellar genes was enhanced in the absence of a functional mcp gene. Furthermore, a lack of the mcp gene also impaired the ability of C. sakazakii to form a biofilm. Our results demonstrate a regulatory role for MCP in diverse biological processes, including the virulence of C. sakazakii ATCC 29544. To the best of our knowledge, this study is the first to elucidate a potential function of a plasmid-encoded MCP homolog in the C. sakazakii sequence type 8 (ST8) lineage.
INTRODUCTION
Cronobacter spp. are Gram-negative, motile, non-spore-forming, facultative anaerobic microorganisms (1, 2) that have been isolated from a wide range of environments, including water, soil, and a variety of fresh and processed foods, such as powdered milk formula for infants (3–9). The organism is considered to be an opportunistic pathogen and has been linked to life-threatening diseases, including necrotizing enterocolitis, septicemia, and meningitis, with a high mortality rate (40 to 80%) in low-birth-weight neonates (7, 9–11). A few reports describe the transmission and virulence of Cronobacter spp.; however, we are still far from completely understanding these mechanisms. Cronobacter spp. can form a biofilm on surfaces, such as glass, stainless steel, polyvinyl chloride, silicone, and enteral feeding tubes, and this biofilm formation could be a vehicle of infection (12–14). The outer membrane proteins OmpA and OmpX from C. sakazakii are reportedly involved in invasion/adhesion to human enterocyte-like Caco-2 and intestinal INT407 epithelial cells (15–17). A LysR-type transcriptional regulator (LTTR) reportedly plays a role in various phenotypes that might be important for the transmission and pathogenesis of C. sakazakii, suggesting a role as a global regulator (18).
Bacterial plasmids are self-replicating and extrachromosomal replicons that can encode a diverse assortment of virulence factors, including antibiotic resistance, toxins, adherence factors, and secretion systems (19–22). Plasmid-borne virulence gene clusters of one species have been found in plasmids of other species or pathogenic groups, suggesting acquisition by horizontal gene transfer (20, 21). Recently, the genomes of two Cronobacter species, Cronobacter sakazakii ATCC BAA-894 and C. turicensis z3032, have been completely sequenced and shown to possess two and three plasmids, respectively (23, 24). In particular, pESA3 (131 kb) of C. sakazakii ATCC BAA-894 (23) and pCTU1 (138 kb) of C. turicensis z3032 (24) were found to be closely related. Franco et al. reported that 97% of 220 Cronobacter species isolates had a homologous RepFIB plasmid, and these two plasmids contain a single RepFIB-like origin of the replication gene repA and encode common virulence factors, an aerobactin-like siderophore and an ABC ferric-iron transporter (eitABCE) (25). pESA3 also encodes an outer membrane protease shown to provide serum resistance to C. sakazakii BAA-894 and enhance host invasion (26). Likewise, C. sakazakii 680 and C. sakazakii ATCC 29544, which belong to the ST8 lineage, reportedly contain a pESA3/pCTU1-like plasmid according to a comparative analysis (23, 27).
Methyl-accepting chemotaxis proteins (MCPs) mediate many of the chemotactic behaviors of bacteria and archaea. Bacteria respond to various environmental signals (28–30) that activate the corresponding MCPs, such as Tar (taxis toward aspartate and maltose, away from nickel and cobalt), Tsr (taxis toward serine, away from leucine, indole, and weak acids), Trg (taxis toward galactose and ribose), and Tap (taxis toward dipeptides) in Escherichia coli (30, 31). The ability of MCPs to adapt to the chemical environment via methylation allows changes in the organism's motility and feedback adaptation (32–34). In addition to chemotaxis, MCPs have been implicated in the virulence of certain pathogens, such as Treponema pallidum, Vibrio cholerae, and Pseudomonas aeruginosa (35–37).
C. sakazakii is known to cause a systemic infection via translocation from the intestinal lumen into the blood circulation by actively invading various epithelial and endothelial cells of human and animal origin (16, 17, 38). While screening the C. sakazakii ATCC 29544 random mutant library for invasion-related virulence factors, we identified a putative MCP that is encoded by a novel plasmid, pCSA2. pCSA2 was completely sequenced and annotated, and the putative MCP in pCSA2 was confirmed to be involved in adhesion and invasion in cultured mammalian cells, organ colonization in rat pups, and the regulation of motility and biofilm formation in C. sakazakii ATCC 29544. Our data imply a regulatory role for MCP in diverse biological processes, including the virulence of the C. sakazakii ST8 lineage, which comprises C. sakazakii ATCC 29544 and 680 (27, 39).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were grown at 37°C in tryptic soy broth (TSB; Difco, Detroit, MI) under aerobic conditions. When necessary, ampicillin, chloramphenicol, and kanamycin were used at 50 μg/ml, 25 μg/ml, and 50 μg/ml, respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| C. sakazakii | ||
| ATCC 29544 | Wild type | ATCC |
| HR101 | mcp::Kmr | This study |
| HR102 | HR101 with pPMCP | This study |
| E. coli | ||
| DH5α | supE44 hsdR17 recA1 gyrA96 thi-1 relA1 | 68 |
| EC100DTM | pir+ | Epicentre |
| Plasmids | ||
| pACYC184 | repp15A Cmr Tetr | 43 |
| pKD13 | repR6Kγ Apr -FRT Kmr -FRT | 42 |
| pKD46 | reppSC101(Ts) Apr ParaBAD γ β exo | 42 |
| pPMCP | pACYC184-mcp | This study |
Random mutagenesis and screening.
Random mutagenesis was performed using the EZ-Tn5pMOD-3<R6Kγori/MCS> transposon system (Epicentre, Madison, WI) according to the manufacturer's instructions. Briefly, the transposon construct was released by restriction digestion with PvuII and then electroporated (1.8 kV) (MicroPulser; Bio-Rad, Hercules, CA) into competent C. sakazakii ATCC 29544. The transformants were selected on tryptic soy agar (TSA; Difco) plates containing kanamycin (50 μg/ml). The resulting colonies were individually cultured and stored at −80°C in TSB containing 15% (vol/vol) glycerol.
Determination of the transposon insertion site.
To locate the transposon insertion site, genomic DNA was isolated from candidate clones that were defective in invasion (see below for invasion assay). After the self-ligation of restriction enzyme-digested DNA according to the manufacturer's protocol (Epicentre), the ligation mixture was electroporated into EC100D pir+, and the transformants were rescued on TSA containing kanamycin (50 μg/ml). The self-ligated vector was recovered by using a plasmid DNA purification kit (DNA-spin; INtRON, South Korea) and sequenced with Tn5-specific primers provided by the manufacturer (pMOD<MCS> forward sequencing primer and pMOD<MCS> reverse sequencing primer).
Complete nucleotide sequencing and bioinformatics.
C. sakazakii ATCC 29544 plasmid DNA was prepared using a plasmid DNA purification kit (DNA-spin; INtRON). The DNA sequence information obtained from transposon insertion site identification was used for primer walking to complete the whole sequence of the plasmid. Primer walking was performed by Macrogen, South Korea. The complete genome of the plasmid was assembled using SeqMan II sequence analysis software (DNASTAR Inc., Madison, WI). The open reading frames (ORFs) were identified with the ORF Finder at the National Center of Bioinformatics site (http://www.ncbi.nlm.nih.gov/projects/gorf/) and GeneMark.hmm prokaryotic (http://exon.gatech.edu/GeneMark/gmhmmp.cgi). The functional analyses of ORFs were conducted using BLASTP (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome) and InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan5/) (40, 41).
Cell culture.
Human enterocyte-like epithelial Caco-2 (ATCC, Manassas, VA) cells were maintained in Eagle's minimum essential medium (EMEM with l-glutamine; ATCC) containing 10% fetal bovine serum (FBS; Invitrogen). Trypsin-treated cells were seeded (approximately 5 × 104 cells per well) into 24-well tissue culture plates (TPP, Switzerland) and grown at 37°C under 5% CO2. The medium was replaced every 2 days. The cell viability was determined with trypan blue staining.
Site-specific mutagenesis of C. sakazakii ATCC 29544.
The one-step gene inactivation method was used to replace the mcp gene in pCSA2 with the kanamycin resistance (Kmr) cassette (42). The Kmr cassette from plasmid pKD13 (42) was amplified using primers mcp-lamb-F and mcp-lamb-R. Primer mcp-lamb-F (5′-ACCATGGCAGTACTTACATTATCCGGGCAATCTGATTCATTGTAGGCTGGAGCTGCTTCG-3′) carries the sequence immediately upstream of the start codon of the mcp gene, followed by the priming site 1 sequence (underlined) of pKD13 (42). Primer mcp-lamb-R (5′-ATCTGTGATGTTCGACAGATATGGGGATATGCACCATGGCATTCCGGGGATCCGTCGACC-3′) harbors the sequence immediately downstream of the stop codon of the mcp gene linked to the priming site 4 sequence (underlined) of pKD13 (42). The resulting PCR product was transformed into C. sakazakii ATCC 29544 and selected for kanamycin resistance. The insertion of the Kmr cassette into the corresponding gene was verified by colony PCR using primers K1 (5′-CAGTCATAGCCGAATAGCCT-3′), mcp-confirm-F (5′-GGTCACCACCATCGTATATTCT-3′), and mcp-confirm-R (5′-GATAAGGCTACACTGAAAGGAC-3′).
Construction of the complementation strain.
The plasmid pPMCP, which contains the MCP coding sequence and its own promoter, was constructed to complement the mcp mutant. The mcp gene was amplified by PCR using the primers mcp-pACYC-F (5′-GAGTGTTTTCCGGATCCCGGAT-3′) and mcp-pACYC-R (5′-GATGGTCGACAGATATGGGGATAT-3′) (underlining indicates restriction enzyme sites BamHI and SalI, repectively) and C. sakazakii ATCC 29544 genomic DNA as a template. The product was introduced between the BamHI and SalI restriction sites of pACYC184 (43). The sequence of the mcp coding region in the recombinant plasmid was confirmed by nucleotide sequencing (Macrogen, South Korea).
Invasion assay.
The invasion assay was conducted as described previously (18), with modifications. Caco-2 cells were grown in EMEM supplemented with 10% FBS. Prior to bacterial infection, a monolayer of 2 × 105 Caco-2 cells was prepared in a 24-well tissue culture plate. Bacteria were prepared by transferring a 1% inoculum from an overnight culture into fresh, prewarmed TSB and incubating the resultant mixture for 3 h (optical density [OD] = 1.5). The bacterial cells were collected by centrifugation (at 10,000 × g for 3 min at 4°C), washed with phosphate-buffered saline (PBS; pH 7.4), resuspended in EMEM with 10% FBS, and then added onto the cell monolayer at a multiplicity of infection (MOI) of 100. After a 1.5-h incubation, the wells were washed three times with prewarmed PBS to remove extracellular bacteria and then incubated for 1.5 h with the prewarmed medium supplemented with 100 μg/ml of gentamicin to kill extracellular bacteria. Subsequently, the wells were washed three times with PBS, lysed in 1% Triton X-100 for 30 min, and then serially diluted in PBS. A dilution of the suspension was plated on TSA medium to enumerate the CFU.
Adhesion assay.
To assess the adhesive ability, the epithelial cells were treated with 0.8 μg/ml of cytochalasin D (CD; Sigma) for 30 min to inhibit the internalization of bacteria (44, 45). The internalization of wild-type (WT) C. sakazakii into Caco-2 cells was inhibited by approximately 70% in the presence of 0.8 μg/ml of cytochalasin D. Prior to bacterial infection, the wells were washed with PBS, and fresh EMEM was added. Subsequently, C. sakazakii, which was prepared in a manner similar to that for the invasion assay, was applied to the Caco-2 cell monolayer at an MOI of 100 and incubated for 45 min. The plates were washed three times with PBS, lysed in 1% Triton X-100, and then serially diluted in PBS. A dilution of the suspensions was plated on TSA medium to enumerate the CFU of adhesive bacteria.
In vivo rat pup virulence assay by CI analysis.
Bacterial cells grown for 3 h were pelleted, washed, and resuspended in PBS. Three-day-old Sprague-Dawley female rat pups were used to assess the virulence of the C. sakazakii WT and HR101 strains. A mixed inoculum of 5 × 109 CFU of WT ATCC 29544 and HR101 in 50 μl of PBS was administered orally to groups of rat pups (5 rats/group). To analyze bacterial colonization in organs, the rat pups were sacrificed 20 h after infection, and the spleen and liver were aseptically removed. The organs were homogenized in 1 ml of ice-cold PBS and serially diluted. The bacterial loads were determined by plating the diluents on TSA plates in the presence or absence of kanamycin. The results are presented as competitive index (CI) values, which were calculated as (mutantoutput/wild typeoutput)/(mutantinput/wild typeinput).
Motility assay.
A 1-μl aliquot of a subculture grown for 3 h in TSB was spotted in the middle of a swim plate (TSA, 0.3% agar) and allowed to dry for 1 h at room temperature. The plates were incubated at 37°C for 8 h.
RNA isolation and qRT-PCR.
To extract RNA from C. sakazakii, bacteria were grown at 37°C for 3 h in TSB. RNA was extracted using an RNeasy minikit (Qiagen), followed by treatment with RNase-free DNase (Ambion). cDNA was synthesized using Omnitranscript reverse transcription (RT) reagents (Qiagen) and random hexamers (Invitrogen) and quantified using 2× iQ SYBR green Supermix (Bio-Rad). The real-time amplification of the PCR products was performed using the iCycler iQ real-time detection system (Bio-Rad). The calculated threshold cycle (CT) corresponding to a target gene was normalized to the CT of the control gene coding the 16S rRNA (46). The primers were designed using a PCR primer design tool, Primer3 Plus. The sequences of the primers used in the quantitative reverse transcription-PCR (qRT-PCR) analysis are listed in Table 2.
TABLE 2.
Primers used in qRT-PCR analysis
| Primer | Target gene | Sequence (5′ to 3′) |
|---|---|---|
| fliA-RT-F | fliA | GCAGGAACTGGGACGTAACG |
| fliA-RT-R | fliA | GTGTCGAGCAACATCTGACGAT |
| fliC-RT-F | fliC | CGTATCGCTGGTGGTGCTAA |
| fliC-RT-R | fliC | CAGCGCCAACCTGAATTTTC |
| flhD-RT-F | flhD | AAGCGTCTGCGATGTTTCG |
| flhD-RT-R | flhD | CAGCCAGTTTCACCATTTGC |
| flhC-RT-F | flhC | GCAACTTAGCCGCGGTAGAC |
| flhC-RT-R | flhC | TGAACCAGTCCGTGGAAAAGG |
| control-RT-F | 16S rRNA | GGGCCTCATGCCATCAGAT |
| control-RT-R | 16S rRNA | TCTCAGACCAGCTAGGGATCGT |
Biofilm assay.
The experiment was performed as previously described (18), with modifications. C. sakazakii was inoculated into 3 ml of TSB and incubated at 37°C with aeration until the cell density reached 2.5 × 108 CFU/ml. The culture was diluted 1:100 in TSB, and 500-μl portions were loaded in triplicate into a 24-well polystyrene plate (SPL Life Sciences, South Korea) and incubated at 37°C for 48 h without shaking. To fix the biofilm, 100 μl of 99% methanol was added for 15 min, the supernatants were removed, and the plates were air dried. Subsequently, 500 μl of crystal violet (CV) solution was added. After 20 min, the excess CV was removed, and the plates were washed with PBS. Finally, the bound CV was released by adding 250 μl of 95% ethanol (Merck). The absorbance was measured at 570 nm using a Sunrise basic microplate reader (Tecan, Austria).
Statistical analysis.
Statistical analyses were conducted using the GraphPad Prism program (version 5.0). All results were analyzed with the unpaired t test. The data are represented as the means ± standard deviations. A P value of <0.05 was considered statistically significant.
Ethics statement.
This study was carried out according to the recommended protocol for the care and use of laboratory animals from the Institute of Laboratory Animal Resources at Seoul National University, which is based on the Korean Animal Protection Law and Korea Food and Drug Administration regulations on laboratory animals. The protocol was approved by the Committee on the Ethics of Animal Experiments of Seoul National University (Institutional Animal Care and Use Committee permit number SNU-130214-1-3).
Nucleotide sequence accession number.
The GenBank accession number for the complete genome sequence and annotation information for pCSA2 is KC663407.
RESULTS AND DISCUSSION
A plasmid-borne gene discovered in an invasion-attenuated mutant.
To identify genes related to the virulence of C. sakazakii, a transposon-mediated random mutant library was constructed in C. sakazakii ATCC 29544, and an invasion-defective clone was screened. Three hundred clones were screened with an invasion assay, and one clone showing the most defective invasion ability was selected and analyzed further. After plasmid recovery and sequencing of the boundary region between the transposon and the C. sakazakii genome, we found that one mutant had the transposon inserted into an unknown region of DNA. A nucleotide BLAST search of the transposon-flanking region showed no homology with Cronobacter spp. but identified part of the mobA gene, which is related to plasmid conjugation (47). This finding suggested that this mutant contained a transposon in a novel plasmid of C. sakazakii ATCC 29544.
To confirm the presence of the boundary region sequence in the plasmid of C. sakazakii ATCC 29544, primers were designed using the boundary sequence from the transposon insertion mutant. The expected size of the PCR amplicon was identified only in the plasmid fraction (data not shown). Taken together, these data implied that C. sakazakii ATCC 29544 contains an unreported plasmid that might be related to its virulence.
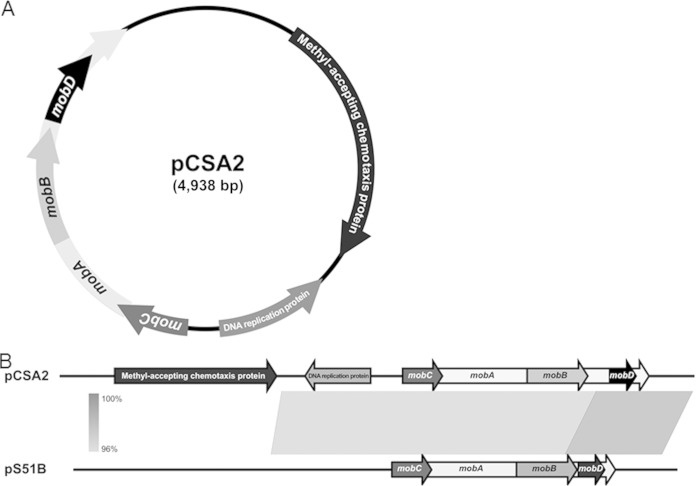
pCSA2 is a novel plasmid in C. sakazakii ATCC 29544 that contains 6 open reading frames.
We obtained the complete plasmid sequence using primer walking and named it pCSA2, which stands for plasmid of C. sakazakii ATCC 29544. The pCSA2 comprises 4,938 bp with an overall G+C content of 54.88%. Six ORFs were identified on pCSA2: four genes predicted to encode relaxases (mobA, mobB, mobC, and mobD), a gene encoding a putative DNA replication protein, and a gene encoding a putative methyl-accepting chemotaxis protein (MCP) (Fig. 1A). We attempted to determine the plasmid profiles of C. sakazakii ATCC 29544 with a repA-targeted PCR assay (25) and found that C. sakazakii ATCC 29544 was PCR positive for pCTU3 and pESA3/pCTU1 but not pESA2/pCTU2 (data not shown). To corroborate the PCR result, the whole genome of C. sakazakii ATCC 29544 was sequenced and compared with that of published plasmids. The resultant comparison of the DNA sequence suggests that C. sakazakii ATCC 29544 contains pCTU3-like, pESA3/pCTU1-like, and pCSA2 sequences (unpublished data). The DNA sequences of the region containing the genes for relaxase and the DNA replication protein are highly similar (97% identities) to those of the Enterobacter cloacae plasmid pS51B, which belongs to the ColE1 superfamily of mobilizable plasmids commonly detected in Enterobacteriaceae (Fig. 1B) (48). A relaxase is a single-stranded DNA transesterase enzyme produced by some prokaryotes and viruses (47). Relaxases are responsible for site- and strand-specific nicks in double-stranded DNA. Mob relaxases nick at the origin of transfer (oriT) to initiate the process of DNA mobilization and transfer known as bacterial conjugation (49). Therefore, the four mob genes might play a role in the conjugation of pCSA2. The DNA replication protein is required for the replication of the plasmid (50).
FIG 1.
Identification of pCSA2 and construction of the mcp deletion mutant by lambda recombination. (A) Schematic representation of pCSA2 in C. sakazakii ATCC 29544. The transposon (Tn5) insertion is designated. (B) Linear comparison diagrams showing BLAST matches between pCSA2 and pS51B, obtained from Easyfig (version 2.1).
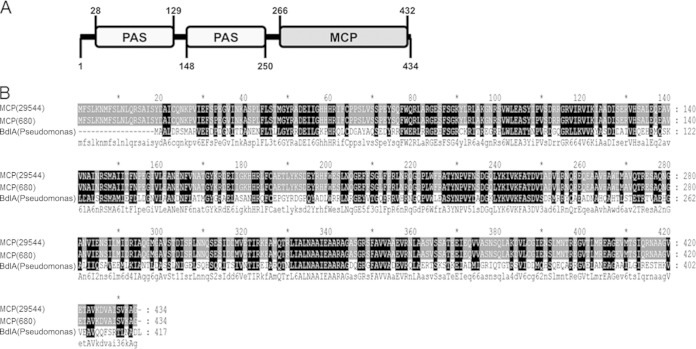
The putative MCP-encoding gene lacked identity to any other gene in C. sakazakii strains except for a recently sequenced C. sakazakii 680 genome (27), which contains a gene with 100% identical DNA sequences. The deduced protein of the putative MCP gene contains an MCP domain and two sensory PAS (Per-Arnt-Sim sensory) domains and high sequence similarity with the biofilm dispersion protein BdlA of Pseudomonas aeruginosa (Fig. 2). Therefore, we named this gene mcp (for methyl-accepting chemotaxis protein). MCP domains share a similar topology and signaling mechanisms. MCPs either bind ligands directly or interact with ligand-binding proteins, transducing the signal to downstream signaling proteins in the cytoplasm (30, 31). The PAS domain is responsible for sensing the input signal and protein-protein interaction. PAS domains have been implicated in diverse biological processes, including the global regulation of metabolism, nitrogen fixation, aerotaxis, hypoxia responses, and ion channel function, in both prokaryotes and eukaryotes (51–54). C. sakazakii is likely to encounter numerous suboptimal conditions during its transition from the environment to the host (55). Therefore, elucidating the mechanisms used by C. sakazakii to regulate environmental signals will be important to understand the pathogenesis of C. sakazakii ATCC 29544. For this, the mcp gene was a reasonable target for further study.
FIG 2.
Deduced sequence analysis of MCP. (A) Domain structures of MCP. PAS, Per-Arnt-Sim sensory domain; MCP, methyl-accepting chemotaxis protein domain. (B) Alignment of amino acid sequences of MCP of C. sakazakii ATCC 29544, MCP of C. sakazakii 680 (ZP_19182295.1), and BdlA of P. aeruginosa PAO1 (NP250114.1) by Clustal Omega (version 1.2.1) and Genedoc. Capital letters in the consensus sequence indicate conserved amino acids appearing in all aligned sequences, and lowercase letters indicate conserved amino acids appearing in at least two sequences.
Plasmid-encoded putative MCP affects adhesion/invasion.
To explore the ability of the mcp gene to contribute to C. sakazakii ATCC 29544 virulence, we constructed a strain in which the entire mcp gene was replaced with a gene encoding kanamycin resistance using lambda red recombination (data not shown). Because the mcp gene is carried in the plasmid, we did not remove the inserted Kmr cassette for the maintenance of the plasmid. Due to the plasmid copy number, we subcultured the mutant on TSA plates containing kanamycin until the mcp PCR product was no longer detected (data not shown). Removing the mcp gene from the plasmid did not result in a significant growth defect compared to growth of the wild-type strain in TSB media (data not shown). After constructing the mcp mutant, the gentamicin protection assay confirmed the observations in the random mutant library screening. The invasion rate of the mutant was significantly lower than that of the WT (approximately 20%, compared to 100%) in the invasion assay (Fig. 3A). This result was comparable to the library screening results. The phenotypic defect of the mcp mutant was indeed due to mcp function, because the expression of the mcp gene from a complement strain enabled the mcp mutant to invade Caco-2 cells at a level similar to that of the wild-type strain (Fig. 3A).
FIG 3.
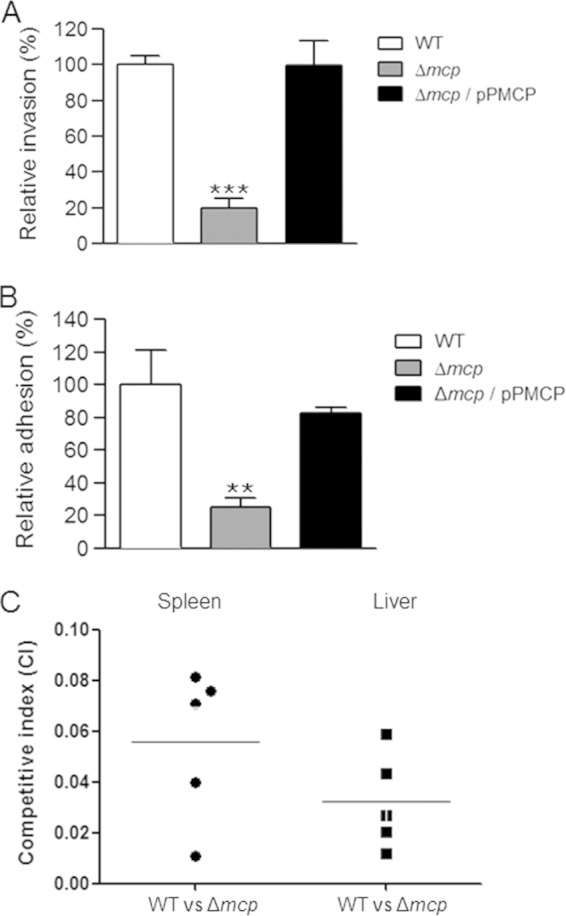
Contribution of the mcp gene to C. sakazakii virulence. (A) Caco-2 epithelial cells were infected with the wild-type (WT) strain (ATCC 29544), the mcp deletion mutant (HR101), or strain HR101 harboring the pPMCP plasmid (Δmcp/pPMCP). The numbers of intracellular bacteria were determined 1.5 h after infection using the gentamicin protection assay. The means and standard deviations from at least three independent experiments are shown. Triple asterisks indicate that the numbers of bacteria were significantly different (P < 0.001) from those of the WT strain. (B) CD-pretreated Caco-2 epithelial cells were infected with C. sakazakii strains similar to those used in the invasion assay. The numbers of intracellular bacteria were determined 45 min after infection without the use of gentamicin. The means and standard deviations from at least three independent experiments are shown. Double asterisks indicate that the numbers of bacteria were significantly different (P < 0.01) from those of the WT strain. (C) Groups of SD rats (5 rats/group) were infected orally with a mixed inoculum of approximately 5 × 109 CFU of the WT and mutant strain. Twenty hours after infection, the numbers of bacteria in the liver and spleen were determined. The competitive index (CI) values were calculated as (mutantoutput/wild typeoutput)/(mutantinput/wild typeinput); thus, a competitive index of <1.0 indicates a strain with a competitive disadvantage.
C. sakazakii needs to bind to the surfaces of epithelial cells to successfully invade them (15). For the adhesion assay, we pretreated Caco-2 cells with cytochalasin D (CD), an agent that causes microfilament depolymerization in eukaryotic cells and thus inhibits C. sakazakii invasion (17, 44). Only 25% of mcp mutants were recovered from the adhesion assay, compared to 100% for the WT (Fig. 3B). Again, the complementary plasmid expressing mcp restored the adhesive defect of the mcp mutant (Fig. 3B). These results indicate that the MCP encoded in pCSA2 is important for the infection of C. sakazakii ATCC 29544.
C. sakazakii lacking a plasmid-encoded putative MCP showed attenuated virulence in rat pups.
The effects of the mcp deletion mutation on C. sakazakii virulence were further analyzed using a newborn rat model (56, 57). A comparison of the CI values of the mcp deletion mutant to those of the wild type revealed an approximately 100-fold-reduced ability to translocate into deep organs (the liver and spleen) in the mcp deletion mutant 20 h after infection in both the livers and spleens of rat pups (Fig. 3C). Taken together, these results suggest that the putative MCP plays a crucial role in the pathogenesis of C. sakazakii ATCC 29544, which was originally isolated from throat culture of a patient with whooping cough (2).
Plasmid-borne MCP regulates the motility of C. sakazakii.
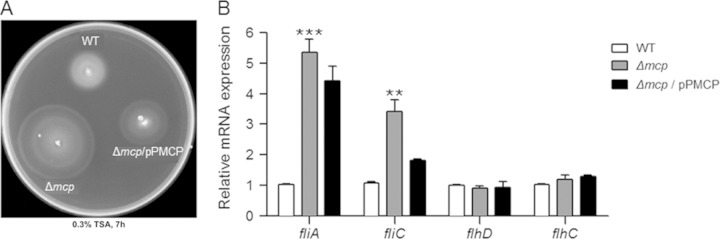
Because mcp is a putative MCP-coding gene and chemotactic regulation in bacteria results in general changes in flagellar rotation (28), we assessed the motility of the WT and the mcp mutant. The wild-type strain was motile but showed low motility (∼15.7 mm in diameter) under standard assay conditions. Conversely, the mcp mutant was hypermotile (∼31.5 mm) (Fig. 4A). Introducing the plasmid expressing the mcp gene reduced the motility (∼17.3 mm) to close to that of the wild-type strain, suggesting that this gene is related to flagellar regulation.
FIG 4.
Effect of the mcp gene on the regulation of motility in C. sakazakii ATCC 29544. (A) A 1-μl aliquot of a subculture was spotted onto the middle of a swim plate (TSA, 0.3% agar). (B) The transcription levels of the fliA, fliC, flhD, and flhC genes in C. sakazakii were determined via qRT-PCR. The bacterial RNA was isolated from the wild-type (WT) strain (ATCC 29544), the mcp deletion mutant (HR101), or strain HR101 harboring the pPMCP plasmid (Δmcp/pPMCP). To obtain the relative mRNA expression values on the y axis, the mRNA level of each gene was divided by the mRNA level of the 16S rRNA-coding gene. The mRNA expression values were further normalized by the transcription levels displayed by the wild-type strain. The means and standard deviations from three independent experiments are shown. Asterisks indicate significant differences (***, P < 0.001; **, P < 0.01).
Next, we evaluated the expression level of several flagellar biosynthesis-related genes in the mcp mutant and wild-type strain by qRT-PCR. We selected four genes, flhD, flhC, fliA, and fliC, that represent the regulation of flagellar assembly (58). These genes from strains BAA-894 and 680 have more than 99% identity except for fliA, which has 92% identity. Due to a lack of information on the genome sequence of C. sakazakii ATCC 29544, we determined the sequences of these genes from the genome sequence data of C. sakazakii BAA-894 and then confirmed the existence of these genes in ATCC 29544, with high sequence identities (data not shown). Interestingly, the mRNA levels of the fliA and fliC genes in the mcp deletion mutant increased ∼5- and ∼3-fold, respectively, compared with the wild-type strain. The complementary plasmid reduced the expression of both fliA and fliC; however, the expression level was not as low as that of the wild-type strain (Fig. 4B). This result might be due to the low copy number of the backbone plasmid pACYC184 (43) in the complementary plasmid, pPMCP. The fliA gene encodes an alternative sigma factor which is responsible for the transcription of class III flagellar genes, including the filament structure genes and the genes for the chemosensory pathway (58, 59). Because the fliC gene is one of the class III flagellar genes (58), the enhanced expression of fliC might be due to the large amount of FliA. These regulations corresponded to the increase in the motility of the mcp mutant. Moreover, the expression levels of two flagellar master regulator genes, flhD and flhC, did not change in the mcp mutant (Fig. 4A), suggesting that MCP affects the expression of class III flagellar genes via fliA.
Motility is a well-known virulence factor in many pathogenic bacteria (60, 61). The motility phenotype is reportedly coupled to the expression of multiple virulence factors, and the negative effects of hypermotility on virulence have been reported for Vibrio cholerae (62, 63) and Ralstonia solanacearum (64). In Vibrio cholerae, hypermotility caused poorer toxin production and defects in colonization in vivo (63, 64). Moreover, the hyperflagellated strain showed a lower ability to form biofilm in Ralstonia solanacearum. Because the attachment of C. sakazakii to the cell surface is a critical step for host invasion (17), the disruption of the adherence of C. sakazakii ATCC 29544 to the host cell (Fig. 3B) due to the hypermotility of the mcp mutant may have caused the colonization defect in vivo, as shown in Fig. 3C.
Biofilm formation is affected by the plasmid-encoded putative MCP.
In the BLAST results, the MCP of C. sakazakii ATCC 29544 showed 63% similarity with the biofilm dispersion protein BdlA of P. aeruginosa (Fig. 2B). In Pseudomonas aeruginosa, BdlA, a chemotaxis transducer protein, is essential for biofilm dispersion (65–67). According to the proposed model of BdlA regulation in P. aeruginosa biofilm dispersion, an active form of BdlA is required for the transition from surface-attached biofilm to the motile lifestyle, and BdlA is intact but inactive under planktonic conditions (66). Therefore, we hypothesized that the deletion of mcp might affect biofilm formation in C. sakazakii. As expected, the mcp deletion mutant resulted in an ∼2-fold reduction of biofilm formation (Fig. 5). The expression of the mcp gene from a complementary plasmid enabled the mcp deletion mutant to form biofilm (∼1.6-fold increase compared with the mutant), albeit at a lower level than for the wild-type strain (Fig. 5).
FIG 5.
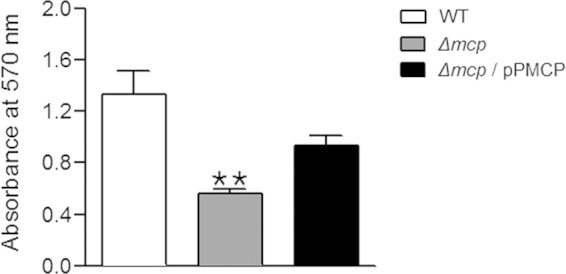
Biofilm formation. The crystal violet biofilm assay was performed in triplicate and repeated three times. The biofilm formation is indicated by the absorbance at 570 nm from the wild-type (WT) strain (ATCC 29544), the mcp deletion mutant (HR101), or strain HR101 harboring the pPMCP plasmid (Δmcp/pPMCP). Double asterisks indicate that the absorbance was significantly different (P < 0.01) from that of the WT strain.
Multilocus sequence typing (MLST) discriminates the Cronobacter genus via a comparison with 7 putative housekeeping genes (atpD, fusA, glnS, gltB, gyrB, infB, and pps), which are necessary for their biological roles in DNA repair, replication, and amino acid biosynthesis (39). Among the many serotypes, ST8 comprises C. sakazakii ATCC 29544 and C. sakazakii 680 (27, 39). Only C. sakazakii 680 matches the MCP sequence 100%, and the MCP sequence is not found in any other lineages, suggesting that the MCP association with virulence is likely specific to the ST8 lineage. However, we demonstrated that the putative MCP encoded in pCSA2 is important for the invasion/adhesion and colonization of C. sakazakii ATCC 29544. MCP also regulated two other phenotypes, motility and biofilm formation. These diverse effects of putative MCP should be further studied, especially for the global regulations of C. sakazakii ATCC 29544.
ACKNOWLEDGMENT
This research was supported by a grant (14162MFDS972) from the Ministry of Food and Drug Safety, South Korea.
REFERENCES
- 1.Iversen C, Mullane N, McCardell B, Tall BD, Lehner A, Fanning S, Stephan R, Joosten H. 2008. Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov., comb. nov., Cronobacter malonaticus sp. nov, Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. nov., Cronobacter dublinensis subsp. lausannensis subsp. nov. and Cronobacter dublinensis subsp. lactaridi subsp. nov. Int J Syst Evol Microbiol 58:1442–1447. doi: 10.1099/ijs.0.65577-0. [DOI] [PubMed] [Google Scholar]
- 2.Farmer JJ, Asbury MA, Hickman FW, Brenner DJ. 1980. Enterobacter sakazakii—a new species of Enterobacteriaceae isolated from clinical specimens. Int J Syst Bacteriol 30:569–584. doi: 10.1099/00207713-30-3-569. [DOI] [Google Scholar]
- 3.Bar-Oz B, Preminger A, Peleg O, Block C, Arad I. 2001. Enterobacter sakazakii infection in the newborn. Acta Paediatr 90:356–358. doi: 10.1111/j.1651-2227.2001.tb00319.x. [DOI] [PubMed] [Google Scholar]
- 4.Biering G, Karlsson S, Clark NC, Jonsdottir KE, Ludvigsson P, Steingrimsson O. 1989. Three cases of neonatal meningitis caused by Enterobacter sakazakii in powdered milk. J Clin Microbiol 27:2054–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farber JM. 2004. Enterobacter sakazakii—new foods for thought? Lancet 363:5–6. doi: 10.1016/S0140-6736(03)15244-0. [DOI] [PubMed] [Google Scholar]
- 6.Kandhai MC, Reij MW, Gorris LG, Guillaume-Gentil O, van Schothorst M. 2004. Occurrence of Enterobacter sakazakii in food production environments and households. Lancet 363:39–40. doi: 10.1016/S0140-6736(03)15169-0. [DOI] [PubMed] [Google Scholar]
- 7.Bowen AB, Braden CR. 2006. Invasive Enterobacter sakazakii disease in infants. Emerg Infect Dis 12:1185–1189. doi: 10.3201/eid1208.051509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai KK. 2001. Enterobacter sakazakii infections among neonates, infants, children, and adults. Case reports and a review of the literature. Medicine 80:113–122. doi: 10.1097/00005792-200103000-00004. [DOI] [PubMed] [Google Scholar]
- 9.Mullane NR, Iversen C, Healy B, Walsh C, Whyte P, Wall PG, Quinn T, Fanning S. 2007. Enterobacter sakazakii: an emerging bacterial pathogen with implications for infant health. Minerva Pediatr 59:137–148. [PubMed] [Google Scholar]
- 10.Forsythe SJ. 2005. Enterobacter sakazakii and other bacteria in powdered infant milk formula. Matern Child Nutr 1:44–50. doi: 10.1111/j.1740-8709.2004.00008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedemann M. 2009. Epidemiology of invasive neonatal Cronobacter (Enterobacter sakazakii) infections. Eur J Clin Microbiol Infect Dis 28:1297–1304. doi: 10.1007/s10096-009-0779-4. [DOI] [PubMed] [Google Scholar]
- 12.Kim H, Bang J, Beuchat LR, Ryu JH. 2008. Fate of Enterobacter sakazakii attached to or in biofilms on stainless steel upon exposure to various temperatures or relative humidities. J Food Prot 71:940–945. [DOI] [PubMed] [Google Scholar]
- 13.Kim H, Ryu JH, Beuchat LR. 2006. Attachment of and biofilm formation by Enterobacter sakazakii on stainless steel and enteral feeding tubes. Appl Environ Microbiol 72:5846–5856. doi: 10.1128/AEM.00654-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jo SH, Baek SB, Ha JH, Ha SD. 2010. Maturation and survival of Cronobacter biofilms on silicone, polycarbonate, and stainless steel after UV light and ethanol immersion treatments. J Food Prot 73:952–956. [DOI] [PubMed] [Google Scholar]
- 15.Kim K, Kim KP, Choi J, Lim JA, Lee J, Hwang S, Ryu S. 2010. Outer membrane proteins A (OmpA) and X (OmpX) are essential for basolateral invasion of Cronobacter sakazakii. Appl Environ Microbiol 76:5188–5198. doi: 10.1128/AEM.02498-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singamsetty VK, Wang Y, Shimada H, Prasadarao NV. 2008. Outer membrane protein A expression in Enterobacter sakazakii is required to induce microtubule condensation in human brain microvascular endothelial cells for invasion. Microb Pathog 45:181–191. doi: 10.1016/j.micpath.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohan Nair MK, Venkitanarayanan K. 2007. Role of bacterial OmpA and host cytoskeleton in the invasion of human intestinal epithelial cells by Enterobacter sakazakii. Pediatr Res 62:664–669. doi: 10.1203/PDR.0b013e3181587864. [DOI] [PubMed] [Google Scholar]
- 18.Choi Y, Kim KP, Kim K, Choi J, Shin H, Kang DH, Ryu S. 2012. Possible roles of LysR-type transcriptional regulator (LTTR) homolog as a global regulator in Cronobacter sakazakii ATCC 29544. Int J Med Microbiol 302:270–275. doi: 10.1016/j.ijmm.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 19.Ahmer BMM, Tran M, Heffron F. 1999. The virulence plasmid of Salmonella typhimurium is self-transmissible. J Bacteriol 181:1364–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson TJ, Nolan LK. 2009. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol Mol Biol. Rev 73:750–774. doi: 10.1128/MMBR.00015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juhas M, Crook DW, Hood DW. 2008. Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell Microbiol 10:2377–2386. doi: 10.1111/j.1462-5822.2008.01187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Actis LA, Tolmasky ME, Crosa JH. 1999. Bacterial plasmids: replication of extrachromosomal genetic elements encoding resistance to antimicrobial compounds. Front Biosci 4:D43–D62. doi: 10.2741/Actis. [DOI] [PubMed] [Google Scholar]
- 23.Kucerova E, Clifton SW, Xia XQ, Long F, Porwollik S, Fulton L, Fronick C, Minx P, Kyung K, Warren W, Fulton R, Feng D, Wollam A, Shah N, Bhonagiri V, Nash WE, Hallsworth-Pepin K, Wilson RK, McClelland M, Forsythe SJ. 2010. Genome sequence of Cronobacter sakazakii BAA-894 and comparative genomic hybridization analysis with other Cronobacter species. PLoS One 5:e9556. doi: 10.1371/journal.pone.0009556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stephan R, Lehner A, Tischler P, Rattei T. 2011. Complete genome sequence of Cronobacter turicensis LMG 23827, a food-borne pathogen causing deaths in neonates. J Bacteriol 193:309–310. doi: 10.1128/JB.01162-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franco AA, Hu L, Grim CJ, Gopinath G, Sathyamoorthy V, Jarvis KG, Lee C, Sadowski J, Kim J, Kothary MH, McCardell BA, Tall BD. 2011. Characterization of putative virulence genes on the related RepFIB plasmids harbored by Cronobacter spp. Appl Environ Microbiol 77:3255–3267. doi: 10.1128/AEM.03023-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franco AA, Kothary MH, Gopinath G, Jarvis KG, Grim CJ, Hu L, Datta AR, McCardell BA, Tall BD. 2011. Cpa, the outer membrane protease of Cronobacter sakazakii, activates plasminogen and mediates resistance to serum bactericidal activity. Infect Immun 79:1578–1587. doi: 10.1128/IAI.01165-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joseph S, Desai P, Ji Y, Cummings CA, Shih R, Degoricija L, Rico A, Brzoska P, Hamby SE, Masood N, Hariri S, Sonbol H, Chuzhanova N, McClelland M, Furtado MR, Forsythe SJ. 2012. Comparative analysis of genome sequences covering the seven Cronobacter species. PLoS One 7:e49455. doi: 10.1371/journal.pone.0049455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Armitage JP. 1999. Bacterial tactic responses. Adv Microb Physiol 41:229–289. doi: 10.1016/S0065-2911(08)60168-X. [DOI] [PubMed] [Google Scholar]
- 29.Falke JJ, Hazelbauer GL. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci 26:257–265. doi: 10.1016/S0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Derr P, Boder E, Goulian M. 2006. Changing the specificity of a bacterial chemoreceptor. J Mol Biol 355:923–932. doi: 10.1016/j.jmb.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 31.Hazelbauer GL, Engstrom P, Harayama S. 1981. Methyl-accepting chemotaxis protein III and transducer gene trg. J Bacteriol 145:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkinson JS. 2003. Bacterial chemotaxis: a new player in response regulator dephosphorylation. J Bacteriol 185:1492–1494. doi: 10.1128/JB.185.5.1492-1494.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barnakov AN, Barnakova LA, Hazelbauer GL. 1999. Efficient adaptational demethylation of chemoreceptors requires the same enzyme-docking site as efficient methylation. Proc Natl Acad Sci U S A 96:10667–10672. doi: 10.1073/pnas.96.19.10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexander RP, Zhulin IB. 2007. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc Natl Acad Sci U S A 104:2885–2890. doi: 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hagman KE, Porcella SF, Popova TG, Norgard MV. 1997. Evidence for a methyl-accepting chemotaxis protein gene (mcp1) that encodes a putative sensory transducer in virulent Treponema pallidum. Infect Immun 65:1701–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee SH, Butler SM, Camilli A. 2001. Selection for in vivo regulators of bacterial virulence. Proc Natl Acad Sci U S A 98:6889–6894. doi: 10.1073/pnas.111581598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLaughlin HP, Caly DL, McCarthy Y, Ryan RP, Dow JM. 2012. An orphan chemotaxis sensor regulates virulence and antibiotic tolerance in the human pathogen Pseudomonas aeruginosa. PLoS One 7:e42205. doi: 10.1371/journal.pone.0042205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim KP, Loessner MJ. 2008. Enterobacter sakazakii invasion in human intestinal Caco-2 cells requires the host cell cytoskeleton and is enhanced by disruption of tight junction. Infect Immun 76:562–570. doi: 10.1128/IAI.00937-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baldwin A, Loughlin M, Caubilla-Barron J, Kucerova E, Manning G, Dowson C, Forsythe S. 2009. Multilocus sequence typing of Cronobacter sakazakii and Cronobacter malonaticus reveals stable clonal structures with clinical significance which do not correlate with biotypes. BMC Microbiol 9:223. doi: 10.1186/1471-2180-9-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. [DOI] [PubMed] [Google Scholar]
- 41.Zdobnov EM, Apweiler R. 2001. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 42.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang AC, Cohen SN. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol 134:1141–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Q, Zhao WD, Zhang K, Fang WG, Hu Y, Wu SH, Chen YH. 2010. PI3K-dependent host cell actin rearrangements are required for Cronobacter sakazakii invasion of human brain microvascular endothelial cells. Med Microbiol Immunol 199:333–340. doi: 10.1007/s00430-010-0168-8. [DOI] [PubMed] [Google Scholar]
- 45.Stevenson BR, Begg DA. 1994. Concentration-dependent effects of cytochalasin D on tight junctions and actin filaments in MDCK epithelial cells. J Cell Sci 107:367–375. [DOI] [PubMed] [Google Scholar]
- 46.Asakura H, Morita-Ishihara T, Yamamoto S, Igimi S. 2007. Genetic characterization of thermal tolerance in Enterobacter sakazakii. Microbiol Immunol 51:671–677. doi: 10.1111/j.1348-0421.2007.tb03955.x. [DOI] [PubMed] [Google Scholar]
- 47.Dyda F, Hickman AB. 2003. A mob of Reps. Structure 11:1310–1311. doi: 10.1016/j.str.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 48.Gomi K, Takahashi N, Yamaguchi F, Tateishi Y, Yoshida K, Fukuchi K. 2012. Characterization of two mobilizable plasmids isolated from Enterobacter cloacae. Rinsho Byori 60:506–515. [PubMed] [Google Scholar]
- 49.Garcillán-Barcia MP, Francia MV, de la Cruz F. 2009. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev 33:657–687. doi: 10.1111/j.1574-6976.2009.00168.x. [DOI] [PubMed] [Google Scholar]
- 50.del Solar G, Giraldo R, Ruiz-Echevarria MJ, Espinosa M, Diaz-Orejas R. 1998. Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev 62:434–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taylor BL, Zhulin IB. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev 63:479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Repik A, Rebbapragada A, Johnson MS, Haznedar JO, Zhulin IB, Taylor BL. 2000. PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol Microbiol 36:806–816. doi: 10.1046/j.1365-2958.2000.01910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhulin IB, Taylor BL, Dixon R. 1997. PAS domain S-boxes in Archaea, Bacteria and sensors for oxygen and redox. Trends Biochem Sci 22:331–333. doi: 10.1016/S0968-0004(97)01110-9. [DOI] [PubMed] [Google Scholar]
- 54.Fabret C, Feher VA, Hoch JA. 1999. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J Bacteriol 181:1975–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alvarez-Ordóñez A, Begley M, Hill C. 2012. Polymorphisms in rpoS and stress tolerance heterogeneity in natural isolates of Cronobacter sakazakii. Appl Environ Microbiol 78:3975–3984. doi: 10.1128/AEM.07835-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hunter CJ, Singamsetty VK, Chokshi NK, Boyle P, Camerini V, Grishin AV, Upperman JS, Ford HR, Prasadarao NV. 2008. Enterobacter sakazakii enhances epithelial cell injury by inducing apoptosis in a rat model of necrotizing enterocolitis. J Infect Dis 198:586–593. doi: 10.1086/590186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Townsend SM, Hurrell E, Gonzalez-Gomez I, Lowe J, Frye JG, Forsythe S, Badger JL. 2007. Enterobacter sakazakii invades brain capillary endothelial cells, persists in human macrophages influencing cytokine secretion and induces severe brain pathology in the neonatal rat. Microbiology 153:3538–3547. doi: 10.1099/mic.0.2007/009316-0. [DOI] [PubMed] [Google Scholar]
- 58.Chevance FF, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol 6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ohnishi K, Kutsukake K, Suzuki H, Iino T. 1990. Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol Gen Genet 221:139–147. [DOI] [PubMed] [Google Scholar]
- 60.Duan QD, Zhou MX, Zhu LQ, Zhu GQ. 2013. Flagella and bacterial pathogenicity. J Basic Microbiol 53:1–8. doi: 10.1002/jobm.201100335. [DOI] [PubMed] [Google Scholar]
- 61.Josenhans C, Suerbaum S. 2002. The role of motility as a virulence factor in bacteria. Int J Med Microbiol 291:605–614. doi: 10.1078/1438-4221-00173. [DOI] [PubMed] [Google Scholar]
- 62.Faruque SM, Nair GB, Mekalanos JJ. 2004. Genetics of stress adaptation and virulence in toxigenic Vibrio cholerae. DNA Cell Biol 23:723–741. doi: 10.1089/dna.2004.23.723. [DOI] [PubMed] [Google Scholar]
- 63.Gardel CL, Mekalanos JJ. 1996. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect Immun 64:2246–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meng FH, Yao J, Allen C. 2011. A MotN mutant of Ralstonia solanacearum is hypermotile and has reduced virulence. J Bacteriol 193:2477–2486. doi: 10.1128/JB.01360-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morgan R, Kohn S, Hwang SH, Hassett DJ, Sauer K. 2006. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J Bacteriol 188:7335–7343. doi: 10.1128/JB.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Petrova OE, Sauer K. 2012. Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc Natl Acad Sci U S A 109:16690–16695. doi: 10.1073/pnas.1207832109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Petrova OE, Sauer K. 2012. PAS domain residues and prosthetic group involved in BdlA-dependent dispersion response by Pseudomonas aeruginosa biofilms. J Bacteriol 194:5817–5828. doi: 10.1128/JB.00780-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]