Figure 2.
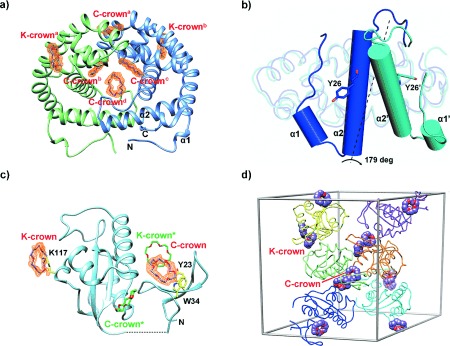
18-Crown-6 binding modes in DMP19 and Pin1-R14A. a) Structure of the DMP19⋅CR dimeric complex. Four C-crowns (C-crowna-d) and two K-crowns (K-crowna and K-crownb), in purple, bind each DMP19 dimer (green and blue). The Fo−Fc omit maps of the CRs (orange) were calculated for each CR and contoured at a 1.5 σ level. α1 and α2 stand for α-helices 1 and 2. b) Comparison of the monomers of the published DMP19 structure (3VJZ, in blue) and the DMP19⋅CR complex (cyan). While 73.6 % of the structure remains practically unchanged (rmsd 0.95 over 108 common Cα atoms), the region between the N-terminus and α-helix 2 changes dramatically, rotating by 179 degrees. The rotated elements are shown as cylinder diagrams, with α1, α2, and Y26 corresponding to 3VJZ, and α1′, α2′, and Y26′ to the DMP19⋅CR complex. c) Ribbon diagram of Pin1R14A⋅CR complex. Two CRs (K- and C-crown) and protein residues interacting with them are shown as stick models (purple, and yellow, respectively). Symmetry related CRs (K- and C-crown*) are shown in green. e) Crystal packing of Pin1R14A and CRs. Six Pin1R14A molecules are shown in different colors in the unit cell. The CRs are presented as sphere diagrams with carbon atoms in purple.
