Figure 1.
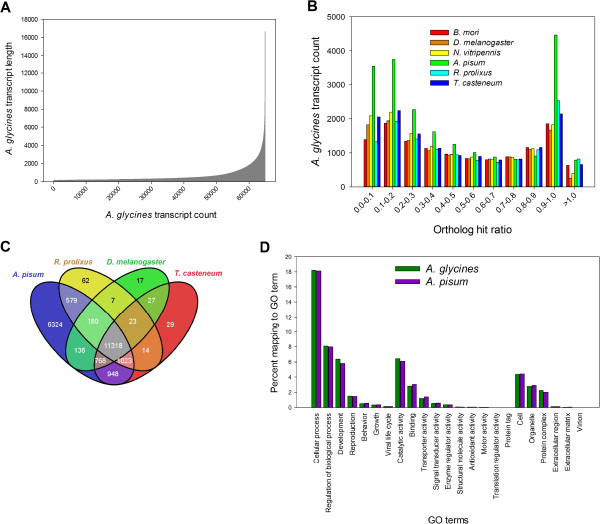
A. glycines transcriptome annotation and comparative genomics. (A) Length distribution of 64,860 contigs in de novo assembly. Individual contigs are ordered on X-axis based on increasing size. (B) Ortholog hit ratio for transcripts calculated after BLASTx searches to genomes of A. pisum, B. mori, D. melanogaster, N. vitripennis, R. prolixus, and T. casteneum . (C) Venn diagram showing the number of transcript contigs with significant matches (unique and common) to genomes of A. pisum, D. melanogaster, R. prolixus, and T. casteneum. Significant matches (e value <1.0E-3) were calculated after pairwise comparisons (BLASTx) to each individual genome. (D) Comparison of GO term mappings distributions of A. glycines and A. pisum that belong to each of the three top-level GO categories (i.e. biological process, molecular function, and cellular component).
