Figure 3.
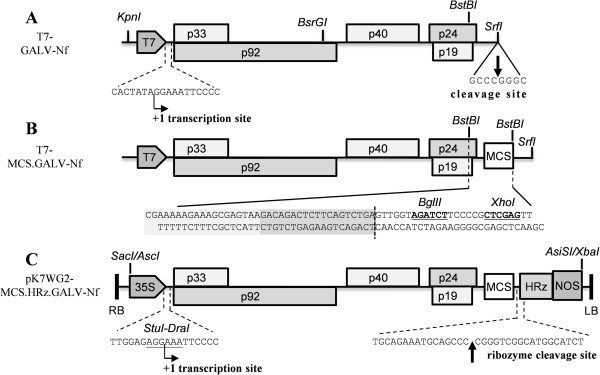
Schematic representation of the infectious GALV-Nf cDNA clones. GALV-Nf sequences under the control of T7 promoter (T7-GALV-Nf and T7-MCS.GALV-Nf) were linearized with SrfI and used for the in vitro production of infectious transcripts (details of T7 transcription site and SrfI cleavage site are shown in panel A). A polylinker was inserted by BstBI cleavage in the T7-GALV-Nf construct obtaining the T7-MCS.GALV-Nf vector (the BglII and XhoI sites of the polylinker are shown in bold and underlined in the sequence reported in panel B). The viral sequence, placed under the control of the CaMV 35S promoter (35S) and the nos terminator (NOS), was introduced (using SacI/AscI and AsiSI/XbaI sites) between the pK7WG2 left and right borders (RB and LB) producing the pK7WG2-MCS.HRz.GALV-Nf binary vector for A. tumefaciens-mediated infection. To design a functional 5′ viral end, the junction between the 35S and the viral sequence was obtained by ligating blunt-end fragments produced by digesting the 35S sequence with StuI and the viral sequence with DraI (details shown in panel C). The sequence of the Hepatitis delta virus antigenomic ribozyme (HRz) was introduced to allow the production of a correct 3′ viral end following ribozyme autocleavage (panel C). T7: T7 promoter; p33 and p92: RNA-dependent RNA polymerases; p40: coat protein (CP); p24: movement protein (MP); p19: silencing suppressor.
