Figure 5.
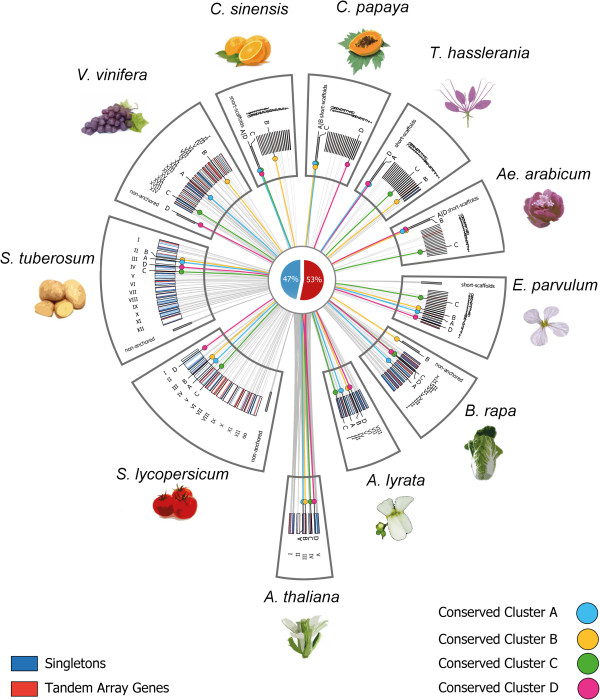
Circos ideogram with 2,363 NB-LRR loci localized on eleven genome annotations. Latin numbers refer to chromosome pseudo-molecules. Loose scaffolds and contigs not anchored to the genome assembly are shown shifted in radius but not in length scale. For genomes without assembly to the chromosome level, the 20 largest scaffolds are displayed and named in in ascending order with Arabic numbers. Beginning at the bottom block in counter-clockwise orientation, shown are (1) Arabidopsis thaliana Col-0, (2) Arabidopsis lyrata, (3) Brassica rapa, (4) Eutrema parvulum, (5) Aethionema arabicum, (6) Tarenaya hasslerania, (7) Carica papaya, (8) Citrus sinensis, (9) Vitis vinifera, (10) Solanum lycopersicum and (11) Solanum tuberosum. Tandem duplicate gene copies are highlighted in red. Singleton genes are highlighted in dark blue. “Conserved Cluster A-D” refers to four distinct A. thaliana NB-LRR loci that have been coded in distant colors for easy visual distinction (A: AT3G14470; B: AT3G50950; C: AT4G33300; D: AT5G17860) including ohnologs in all other ten genomes. For genome assembly versions used in this analysis, see Figure 3. Please note that due to the fragmented assembly status of Nicotiana benthamiana, all scaffolds of this annotation are below visible length threshold.
