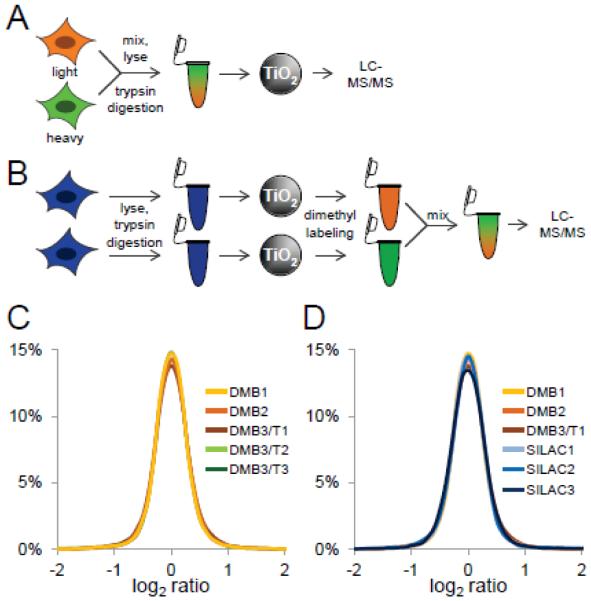
Figure 1. Assessment of post-phosphopeptide enrichment chemical tagging.
A, SILAC scheme. HeLa cells were metabolically labeled with light (orange) and heavy (green) amino acids in cell culture. Labeled cells were mixed, trypsin-digested, phosphopeptides were enriched using titanium dioxide microspheres, and analyzed by LC-MS/MS. B, Reductive dimethyl-labeling scheme. HeLa cells were trypsin digested, and phosphopeptides were enriched using titanium dioxide microspheres. Phosphopeptides were labeled separately using heavy and light reductive dimethyl-labeling chemistry, mixed, and analyzed by LC-MS/MS. C, Log2 ratio distribution of biological (DM1-3) and technical (DMT1-3) replicates of phosphopeptides labeled by reductive dimethyl-labeling chemistry. D, Log2 ratio distribution of biological replicates of phosphopeptides labeled by reductive dimethyl-labeling chemistry (DM1-3) and SILAC (SILAC1-3).