Figure 3.
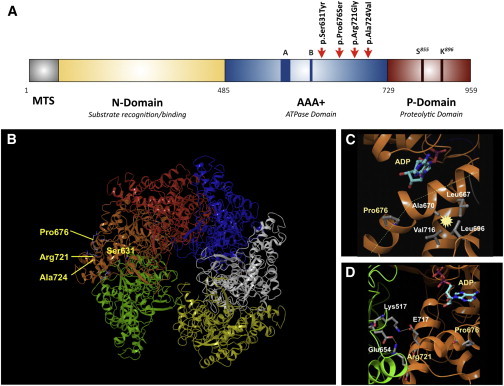
Location of Amino Acids That Are Mutated within Mitochondrial Lon in CODAS Syndrome
(A) Domain structure of the human Lon subunit. Mitochondrial targeting sequence (MTS), substrate recognition/binding (N) domain, ATPase domain (AAA+), and a protease domain (P). Red arrows indicate the position of pathogenic CODAS mutations.
(B) Homology model of human mitochondrial Lon. The model shows the position, within a single Lon subunit (shown in amber), of amino acids that are altered in CODAS syndrome.
(C and D) The positions of amino acids Pro676 and Arg721 (yellow), which are altered in proteins encoded by CODAS homozygous alleles. ADP is shown occupying the ATP- and ADP-binding pocket (a green dashed line represents a proline-induced kink in the helix; a yellow dotted line represents a salt bridge; starbursts represent hydrophobic interactions). (C) The Pro676-induced kink promotes hydrophobic interactions between Ala670, Leu667, Leu696, and Val716. (D) The positions of Arg721 as well as Pro676, Glu717, and the ATP- and ADP-binding site are located on the same Lon subunit (amber). Arg721 and Glu717 form salt bridges with Glu654 and Lys517, respectively, which are located on the adjacent Lon subunit (green).
